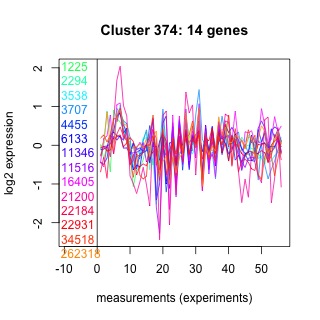
Thaps_hclust_0374 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006342 | chromatin silencing | 0.0129601 | 1 | Thaps_hclust_0374 |
| GO:0016310 | phosphorylation | 0.0186738 | 1 | Thaps_hclust_0374 |
| GO:0006512 | ubiquitin cycle | 0.0509804 | 1 | Thaps_hclust_0374 |
| GO:0006281 | DNA repair | 0.0647398 | 1 | Thaps_hclust_0374 |
| GO:0006464 | protein modification | 0.0823711 | 1 | Thaps_hclust_0374 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.368277 | 1 | Thaps_hclust_0374 |
| GO:0008152 | metabolism | 0.459719 | 1 | Thaps_hclust_0374 |
|
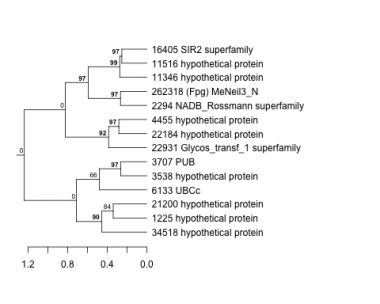
1225 : hypothetical protein |
4455 : hypothetical protein |
16405 : SIR2 superfamily |
22931 : Glycos_transf_1 superfamily |
|
2294 : NADB_Rossmann superfamily |
6133 : UBCc |
21200 : hypothetical protein |
34518 : hypothetical protein |
|
3538 : hypothetical protein |
11346 : hypothetical protein |
22184 : hypothetical protein |
262318 : (Fpg) MeNeil3_N |
|
3707 : PUB |
11516 : hypothetical protein |
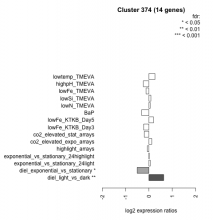
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.634 | 0.00714 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.224 | 0.458 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.199 | 0.425 |
| BaP | BaP | -0.308 | 0.239 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.013 | 0.948 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.197 | 0.463 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.261 | 0.385 |
| highpH_TMEVA | highpH_TMEVA | -0.239 | 0.187 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.123 | 0.646 |
| lowFe_TMEVA | lowFe_TMEVA | -0.122 | 0.769 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.056 | 0.9 |
| lowN_TMEVA | lowN_TMEVA | 0.055 | 0.874 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.502 | 0.0149 |
| lowSi_TMEVA | lowSi_TMEVA | 0.087 | 0.997 |
| highlight_arrays | highlight_arrays | -0.066 | 0.76 |