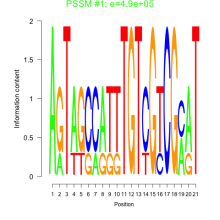
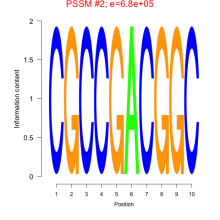
Phatr_bicluster_0085 Residual: 0.45
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0085 | 0.45 | Phaeodactylum tricornutum |
Displaying 1 - 30 of 30
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_1457 nls receptor (ARM)
PHATRDRAFT_14645 iojap-related protein (Oligomerization)
PHATRDRAFT_14849 family protein (BolA superfamily)
PHATRDRAFT_15281 vacuolar transporter (VTC1)
PHATRDRAFT_15595 tryptophanyl-trna synthetase (PRK00927)
PHATRDRAFT_16262 chromatin remodeling factor chrom1 isoform 1 (SNF2_N)
PHATRDRAFT_17029 rna polymerase sigma factor (Sig70-cyanoRpoD)
PHATRDRAFT_17043 drug resistance atpase-1 (drug ra1) abc transporteratp-binding protein (Uup)
PHATRDRAFT_17172 gtp-binding protein (PRK00093)
PHATRDRAFT_18027 delta-1-pyrroline 5-carboxylase synthetase (PLN02418)
PHATRDRAFT_18080 phosphoribosylamidoimidazole-succinocarboxamide synthase (SAICAR_synt_Sc)
PHATRDRAFT_22510 delta 5 fatty acid desaturase (Delta6-FADS-like)
PHATRDRAFT_29004 inositol monophosphatase family protein (IMPase)
PHATRDRAFT_38930 carbon-nitrogen hydrolase (CPA)
PHATRDRAFT_39046 at3g04650 f7o18_13 (NAD_binding_8 superfamily)
PHATRDRAFT_43507 (CRAL_TRIO_2)
PHATRDRAFT_45871 hepatocellular carcinoma-associated antigen 66 (U3_assoc_6 superfamily)
PHATRDRAFT_47239 solute carrier family 34 (sodium phosphate)member 1 (Na_Pi_cotrans)
PHATRDRAFT_47489 intestinal mucin
PHATRDRAFT_47558 elicitin-like protein 6 precursor
PHATRDRAFT_47588 mgc85567 protein
PHATRDRAFT_48958 spou protein (SpoU_methylase)
PHATRDRAFT_49488 cog3781: membrane protein (Bestrophin superfamily)
PHATRDRAFT_50885 glycyl-trna synthetase (PRK04173)
PHATRDRAFT_51214 (gltB)
PHATRDRAFT_51305 carbonic anhydrase family protein (beta_CA_cladeA)
PHATRDRAFT_54958 (SPFH_prohibitin)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006807 | nitrogen compound metabolism | 0.00246558 | 1 | Phatr_bicluster_0085 |
| GO:0015976 | carbon utilization | 0.0108387 | 1 | Phatr_bicluster_0085 |
| GO:0006426 | glycyl-tRNA aminoacylation | 0.0108387 | 1 | Phatr_bicluster_0085 |
| GO:0006436 | tryptophanyl-tRNA aminoacylation | 0.0108387 | 1 | Phatr_bicluster_0085 |
| GO:0006164 | purine nucleotide biosynthesis | 0.0215652 | 1 | Phatr_bicluster_0085 |
| GO:0006561 | proline biosynthesis | 0.0268867 | 1 | Phatr_bicluster_0085 |
| GO:0006537 | glutamate biosynthesis | 0.0268867 | 1 | Phatr_bicluster_0085 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0317216 | 1 | Phatr_bicluster_0085 |
| GO:0006396 | RNA processing | 0.0363639 | 1 | Phatr_bicluster_0085 |
| GO:0006817 | phosphate transport | 0.037447 | 1 | Phatr_bicluster_0085 |

Comments