Phatr_bicluster_0101 Residual: 0.21
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0101 | 0.21 | Phaeodactylum tricornutum |
Displaying 1 - 17 of 17
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10628 polo-like kinase 1 (S_TKc)
PHATRDRAFT_22921 cmp-sialic acid transporter (Nuc_sug_transp)
PHATRDRAFT_37268 (SMC_prok_B)
PHATRDRAFT_43122 protein regulator of cytokinesis 1 isoform 2 (MAP65_ASE1)
PHATRDRAFT_43936 at hook motif familyexpressed (AAA)
PHATRDRAFT_46020 histone h2a (PLN00157)
PHATRDRAFT_47734 proteophosphoglycan ppg4
PHATRDRAFT_48735 chu large protein
PHATRDRAFT_51024 dna-binding horma domain-containing protein (HORMA)
PHATRDRAFT_54677 dpod1_orysj dna polymerase delta catalytic subunit (PTZ00166)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006468 | protein amino acid phosphorylation | 0.262204 | 1 | Phatr_bicluster_0101 |
| GO:0006508 | proteolysis and peptidolysis | 0.328797 | 1 | Phatr_bicluster_0101 |
| GO:0015780 | nucleotide-sugar transport | 0.00689992 | 1 | Phatr_bicluster_0101 |
| GO:0008643 | carbohydrate transport | 0.0273554 | 1 | Phatr_bicluster_0101 |
| GO:0006334 | nucleosome assembly | 0.0273554 | 1 | Phatr_bicluster_0101 |
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 0.0290436 | 1 | Phatr_bicluster_0101 |
| GO:0006979 | response to oxidative stress | 0.0407909 | 1 | Phatr_bicluster_0101 |

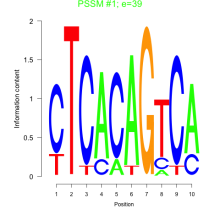

Comments