Phatr_bicluster_0111 Residual: 0.46
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0111 | 0.46 | Phaeodactylum tricornutum |
Displaying 1 - 48 of 48
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_14553 dual-specificity tyrosine--phosphorylation regulated kinaseisoform cra_e (S_TKc)
PHATRDRAFT_15565 rna exonuclease 4 (DnaQ_like_exo superfamily)
PHATRDRAFT_16341 solute carrier family 35 (UAA)
PHATRDRAFT_22559 disease resistance protein
PHATRDRAFT_2593 (PLN00113)
PHATRDRAFT_35692 (DUF501)
PHATRDRAFT_36162 hsf-type dna-binding domain containing protein
PHATRDRAFT_36184 (Periplasmic_Binding_Protein_Type_1 superfamily)
PHATRDRAFT_40792 grip1 associated proteinisoform cra_a
PHATRDRAFT_42434 aaa-type atpase family protein ankyrin repeat family protein (AAA)
PHATRDRAFT_42588 major surface like glycoprotein (Peptidase_M8 superfamily)
PHATRDRAFT_42809 tpr repeat:nb-arc:peptidasecaspase catalytic subunit p20:tetratricopeptide tpr_3:tetratricopeptide tpr_4:tetratricopeptide tpr_4 (TPR_12)
PHATRDRAFT_44023 (DENN)
PHATRDRAFT_44116 tpr repeat:nb-arc:peptidasecaspase catalytic subunit p20:tetratricopeptide tpr_3:tetratricopeptide tpr_4:tetratricopeptide tpr_4 (TPR_12)
PHATRDRAFT_44629 lipopolysaccharide a protein (CAP10 superfamily)
PHATRDRAFT_44975 (Glyco_transf_64 superfamily)
PHATRDRAFT_45748 tkl family protein kinase (S_TKc)
PHATRDRAFT_45751 bnr repeat domain protein
PHATRDRAFT_46087 fibronectin type iii domain protein (Peptidases_S53_like)
PHATRDRAFT_47698 serine threonine protein kinase (PKc)
PHATRDRAFT_48262 peptidase s8 andsedolisin (Peptidases_S8_Subtilisin_subset)
PHATRDRAFT_48623 kinesin motor domain containingexpressed
PHATRDRAFT_49073 (Aa_trans superfamily)
PHATRDRAFT_49102 (TPR_12)
PHATRDRAFT_4937 sh2 domain-containing protein (PKc_like superfamily)
PHATRDRAFT_49426 spore formation protein (AAA)
PHATRDRAFT_49671 ankyrin repeat protein (ANK)
PHATRDRAFT_50632 disease resistance protein
PHATRDRAFT_52304 dot1_gibze histone-lysine n-h3 lysine-79 specific (histone h3-k79 methyltransferase) (h3-k79-hmtase) (DOT1 superfamily)
PHATRDRAFT_52603 epsilon frustilin
PHATRDRAFT_54026 phosphoacetylglucosamine mutase (PLN02895)
PHATRDRAFT_5608 mate efflux family protein (MATE_like superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007155 | cell adhesion | 0.00157814 | 1 | Phatr_bicluster_0111 |
| GO:0006468 | protein amino acid phosphorylation | 0.00738542 | 1 | Phatr_bicluster_0111 |
| GO:0007160 | cell-matrix adhesion | 0.014015 | 1 | Phatr_bicluster_0111 |
| GO:0006334 | nucleosome assembly | 0.0726248 | 1 | Phatr_bicluster_0111 |
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 0.0769938 | 1 | Phatr_bicluster_0111 |
| GO:0006508 | proteolysis and peptidolysis | 0.0841501 | 1 | Phatr_bicluster_0111 |
| GO:0006979 | response to oxidative stress | 0.107035 | 1 | Phatr_bicluster_0111 |
| GO:0006865 | amino acid transport | 0.123783 | 1 | Phatr_bicluster_0111 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.160385 | 1 | Phatr_bicluster_0111 |
| GO:0006512 | ubiquitin cycle | 0.164361 | 1 | Phatr_bicluster_0111 |

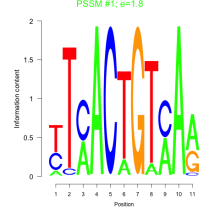

Comments