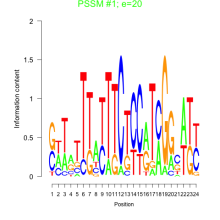
Phatr_bicluster_0114 Residual: 0.31
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0114 | 0.31 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10974 phosphatidylinositol-4-phosphate 5-11335-7537 (MORN superfamily)
PHATRDRAFT_11154 (sufA)
PHATRDRAFT_11271 (Ist1)
PHATRDRAFT_12309 g10 protein (G10)
PHATRDRAFT_13557 2og-fe oxygenase familyexpressed (P4Hc)
PHATRDRAFT_14696 peptidyl-prolyl cis-transcyclophilin type (Pro_isomerase)
PHATRDRAFT_37738 l-a virus gag protein n-acetyltransferase (RimI)
PHATRDRAFT_41220 at5g47860 mca23_20 (DUF1350 superfamily)
PHATRDRAFT_42683 phospholipase c-delta1 (PI-PLCc_eukaryota)
PHATRDRAFT_44433 lysyl hydroxylase
PHATRDRAFT_44454 metallo-beta-lactamase superfamily protein (COG1237 superfamily)
PHATRDRAFT_49521 ctr1-like kinase kinase kinase (S_TKc)
PHATRDRAFT_8996 rac-beta serine threonine protein (STKc_AGC)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006629 | lipid metabolism | 0.0350457 | 1 | Phatr_bicluster_0114 |
| GO:0006468 | protein amino acid phosphorylation | 0.0424353 | 1 | Phatr_bicluster_0114 |
| GO:0019538 | protein metabolism | 0.069012 | 1 | Phatr_bicluster_0114 |
| GO:0007242 | intracellular signaling cascade | 0.0800992 | 1 | Phatr_bicluster_0114 |
| GO:0007165 | signal transduction | 0.0892501 | 1 | Phatr_bicluster_0114 |
| GO:0016567 | protein ubiquitination | 0.13382 | 1 | Phatr_bicluster_0114 |
| GO:0006457 | protein folding | 0.209255 | 1 | Phatr_bicluster_0114 |

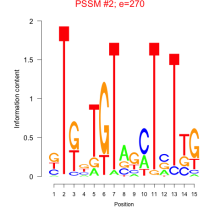
Comments