Phatr_bicluster_0171 Residual: 0.38
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0171 | 0.38 | Phaeodactylum tricornutum |
Displaying 1 - 29 of 29
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10418 aspartate aminotransferase (COG0436)
PHATRDRAFT_1174 udp-glucose:sterol glucosyltransferase (GT1_Gtf_like)
PHATRDRAFT_15102 (COG0390)
PHATRDRAFT_15134 phosphoribulokinase uridine kinase family protein (NK superfamily)
PHATRDRAFT_16932 (FTase)
PHATRDRAFT_2037 ancient conserved domain protein 2 (cyclin m2) (DUF21)
PHATRDRAFT_33116 zifl1 (zinc induced facilitator-like 1) tetracycline:hydrogen antiporter transporter (MFS_1)
PHATRDRAFT_33473 (PHA03247)
PHATRDRAFT_39605 mechanosensitive ion channel domain-containing (MS_channel superfamily)
PHATRDRAFT_40521 vesicle transport v-snare (vesicle soluble nsf attachment protein receptor) (V-SNARE)
PHATRDRAFT_41604 probable cell surface glycoprotein
PHATRDRAFT_41608 redoxin domain protein (PRX_BCP)
PHATRDRAFT_41624 lipase class 3 family protein (Lipase_3)
PHATRDRAFT_47430 rna polymerase ii subunit (RNAP_II_RPB11)
PHATRDRAFT_47924 selected as a weak suppressor of a mutant of the subunit ac40 of dna dependant rna polymerase i and iii
PHATRDRAFT_49805 (RDD)
PHATRDRAFT_50172 chaperone protein (DnaJ-X superfamily)
PHATRDRAFT_50174 rhodanese domain protein (PRK00142)
PHATRDRAFT_5508 kin17 cg5649-pa (Kin17_mid)
PHATRDRAFT_6437 cytochrome c heme lyase (Cyto_heme_lyase superfamily)
PHATRDRAFT_7934 dnajb11 protein (DnaJ)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0030259 | lipid glycosylation | 0.00591934 | 1 | Phatr_bicluster_0171 |
| GO:0006457 | protein folding | 0.0452262 | 1 | Phatr_bicluster_0171 |
| GO:0006629 | lipid metabolism | 0.0521304 | 1 | Phatr_bicluster_0171 |
| GO:0006350 | transcription | 0.0605726 | 1 | Phatr_bicluster_0171 |
| GO:0006979 | response to oxidative stress | 0.0689457 | 1 | Phatr_bicluster_0171 |
| GO:0006886 | intracellular protein transport | 0.112459 | 1 | Phatr_bicluster_0171 |
| GO:0009058 | biosynthesis | 0.125669 | 1 | Phatr_bicluster_0171 |
| GO:0005975 | carbohydrate metabolism | 0.171749 | 1 | Phatr_bicluster_0171 |
| GO:0006810 | transport | 0.440472 | 1 | Phatr_bicluster_0171 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.487367 | 1 | Phatr_bicluster_0171 |

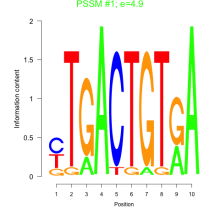
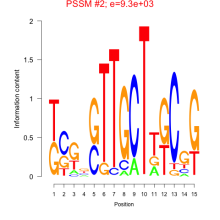
Comments