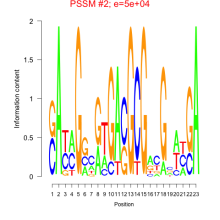
Phatr_bicluster_0190 Residual: 0.25
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0190 | 0.25 | Phaeodactylum tricornutum |
Displaying 1 - 32 of 32
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10824 smt1_orysj cycloartenol-c-24-methyltransferase 1 (24-sterol c-methyltransferase 1) (sterol c-methyltransferase 1) (AdoMet_MTases)
PHATRDRAFT_12233 ubiquinol-cytochrome c reductase iron-sulfurmitochondrial precursor (Rieske_cytochrome_bc1)
PHATRDRAFT_12620 n-ethylmaleimide-sensitive factor nsf sec18p (AAA)
PHATRDRAFT_12989 1 aminotransferase family protein (AAT_I superfamily)
PHATRDRAFT_13034 sideroflexin 5 (Mtc superfamily)
PHATRDRAFT_13123 dehydrogenase kinase (BCDHK_Adom3)
PHATRDRAFT_17967 homolog 1 (FtsJ)
PHATRDRAFT_19699 1-aminocyclopropane-1-carboxylate synthase (AAT_like)
PHATRDRAFT_21420 n-terminal acetyltransferase (NARP1)
PHATRDRAFT_28833 zincste24 homolog (Peptidase_M48)
PHATRDRAFT_32976 cg13865-isoform a (COG0790)
PHATRDRAFT_35336 (DUF2997)
PHATRDRAFT_35647 heme oxygenase (HemeO superfamily)
PHATRDRAFT_42650 (PHA03247)
PHATRDRAFT_45207 fibronectin-binding protein (DUF814 superfamily)
PHATRDRAFT_4550 ac010924_18 ests gb
PHATRDRAFT_45813 pap9_arath probable plastid-lipid-associated proteinchloroplast precursor (fibrillin-9) (PAP_fibrillin superfamily)
PHATRDRAFT_45899 homeodomain protein (Gst)
PHATRDRAFT_46174 (DUF1826 superfamily)
PHATRDRAFT_48613 tpr repeat
PHATRDRAFT_49056 threonine synthase (PRK09225)
PHATRDRAFT_51757 sterol c-22 desaturase -like (p450 superfamily)
PHATRDRAFT_5423 thiamin pyrophosphokinase 1 (PLN02714)
PHATRDRAFT_5762 ac079935_17pyridoxamine 5-phosphate oxidase (PLN03050)
PHATRDRAFT_7294 snat_bovin serotonin n-acetyltransferase (RimI)
PHATRDRAFT_9634 snat_bovin serotonin n-acetyltransferase (RimI)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009229 | thiamin diphosphate biosynthesis | 0.00321067 | 1 | Phatr_bicluster_0190 |
| GO:0006788 | heme oxidation | 0.00960348 | 1 | Phatr_bicluster_0190 |
| GO:0009088 | threonine biosynthesis | 0.00960348 | 1 | Phatr_bicluster_0190 |
| GO:0016310 | phosphorylation | 0.0440926 | 1 | Phatr_bicluster_0190 |
| GO:0006520 | amino acid metabolism | 0.0744808 | 1 | Phatr_bicluster_0190 |
| GO:0006812 | cation transport | 0.0834219 | 1 | Phatr_bicluster_0190 |
| GO:0009058 | biosynthesis | 0.135415 | 1 | Phatr_bicluster_0190 |
| GO:0006468 | protein amino acid phosphorylation | 0.431733 | 1 | Phatr_bicluster_0190 |
| GO:0006810 | transport | 0.466944 | 1 | Phatr_bicluster_0190 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.515177 | 1 | Phatr_bicluster_0190 |

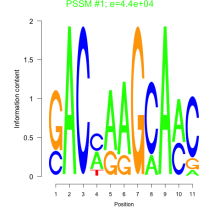
Comments