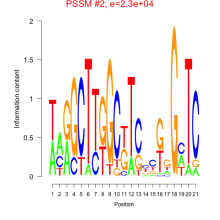
Phatr_bicluster_0211 Residual: 0.25
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0211 | 0.25 | Phaeodactylum tricornutum |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11390 digalactosyldiacylglycerol synthaseexpressed (Glycos_transf_1 superfamily)
PHATRDRAFT_12265 uncharacterized protein family upf0016 containingexpressed (COG2119)
PHATRDRAFT_17335 aldo keto reductase (Aldo_ket_red)
PHATRDRAFT_33977 phosphoglycerate mutase 1 (HP_PGM_like)
PHATRDRAFT_35318 kynurenine 3- (UbiH)
PHATRDRAFT_35638 rhomboid family protein (Rhomboid superfamily)
PHATRDRAFT_43150 oxidoreductase domain protein (MviM)
PHATRDRAFT_43583 (RING)
PHATRDRAFT_43715 rna methylase
PHATRDRAFT_44702 (Amidohydro_2)
PHATRDRAFT_45250 (DUF3321 superfamily)
PHATRDRAFT_45657 (Sad1_UNC superfamily)
PHATRDRAFT_45726 myo-inositol oxygenase (DUF706 superfamily)
PHATRDRAFT_46142 cysteine endopeptidase (Peptidase_C1A)
PHATRDRAFT_46336 binding protein 1 (BRCT)
PHATRDRAFT_46455 at3g61320 t20k12_220 (Bestrophin superfamily)
PHATRDRAFT_46850 crumbs homolog 1isoform cra_b
PHATRDRAFT_49214 unusual protein kinase (AarF)
PHATRDRAFT_7072 isoform cra_b
PHATRDRAFT_9617 tata box binding protein-associated factor (COG1936)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006075 | beta-1,3 glucan biosynthesis | 0.00321067 | 1 | Phatr_bicluster_0211 |
| GO:0008152 | metabolism | 0.0115207 | 1 | Phatr_bicluster_0211 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0347981 | 1 | Phatr_bicluster_0211 |
| GO:0006725 | aromatic compound metabolism | 0.0624342 | 1 | Phatr_bicluster_0211 |
| GO:0006096 | glycolysis | 0.106876 | 1 | Phatr_bicluster_0211 |
| GO:0006508 | proteolysis and peptidolysis | 0.159337 | 1 | Phatr_bicluster_0211 |
| GO:0006118 | electron transport | 0.165064 | 1 | Phatr_bicluster_0211 |
| GO:0016567 | protein ubiquitination | 0.20832 | 1 | Phatr_bicluster_0211 |


Comments