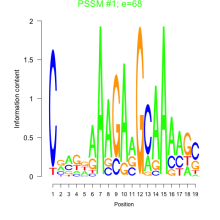
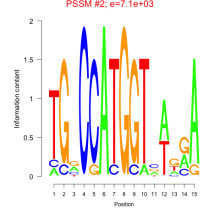
Phatr_bicluster_0239 Residual: 0.32
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0239 | 0.32 | Phaeodactylum tricornutum |
Displaying 1 - 41 of 41
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_16036 protein kinase (PKc_like superfamily)
PHATRDRAFT_16073 rh26_orysj dead-box atp-dependent rna helicase 26 (DEADc)
PHATRDRAFT_22332 htransporter (pgp related) family member (haf-4) (MdlB)
PHATRDRAFT_37957 cyanobacterial globin family protein (globin_like superfamily)
PHATRDRAFT_38038 piggybac transposase uribo2 (DDE_Tnp_1_7 superfamily)
PHATRDRAFT_40303 (DUF1749 superfamily)
PHATRDRAFT_42126 kdel receptor a (ER_lumen_recept superfamily)
PHATRDRAFT_43919 histone deacetylase superfamily (Arginase_HDAC superfamily)
PHATRDRAFT_45387 piggybac transposase uribo2 (DDE_Tnp_1_7 superfamily)
PHATRDRAFT_46983 membrane protein (Lung_7-TM_R superfamily)
PHATRDRAFT_47576 phosphatidylglycerophosphate synthase 1 (PLDc_CDP-OH_P_transf_II_1)
PHATRDRAFT_47789 cortactin-binding protein 2
PHATRDRAFT_47857 mgc80916 protein (Sulfotransfer_2 superfamily)
PHATRDRAFT_47870 lipolytic protein g-d-s-l family (SGNH_hydrolase superfamily)
PHATRDRAFT_48672 (RAP)
PHATRDRAFT_48723 membrane-associated zn-dependent proteases 1 (TIGR00054)
PHATRDRAFT_48755 krox-like protein (TLD)
PHATRDRAFT_48866 ximpact ortholog conserved protein seen in bacteria and eukaryotes (UPF0029)
PHATRDRAFT_48977 mgc81046 protein (KDSR-like_SDR_c)
PHATRDRAFT_49009 pentatricopeptiderepeat-containing protein
PHATRDRAFT_49261 zinc finger constans-like protein (CCT)
PHATRDRAFT_49447 cpi1 (cyclopropyl isomerase)
PHATRDRAFT_49791 integrin alpha beta-propellor repeat protein (Methyltransf_21 superfamily)
PHATRDRAFT_50103 glutamate ornithine acetyltransferase (DUF647 superfamily)
PHATRDRAFT_50162 (DUF647 superfamily)
PHATRDRAFT_50185 pseudouridylate synthase-like 1 (truA)
PHATRDRAFT_50413 (Nucleotid_trans superfamily)
PHATRDRAFT_50417 (Nucleotid_trans superfamily)
PHATRDRAFT_50446 vacuolar protein sorting 54 (Vps54 superfamily)
PHATRDRAFT_52412 chloride channel 7 (Voltage_gated_ClC superfamily)
PHATRDRAFT_54844 n-acetylglucosaminyltransferase i (Glyco_tranf_GTA_type superfamily)
PHATRDRAFT_54869 af214560_1ran gtpase activating protein 2 (LRR_RI superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015671 | oxygen transport | 0.00984608 | 1 | Phatr_bicluster_0239 |
| GO:0006306 | DNA methylation | 0.0483639 | 1 | Phatr_bicluster_0239 |
| GO:0008033 | tRNA processing | 0.0483639 | 1 | Phatr_bicluster_0239 |
| GO:0015031 | protein transport | 0.074057 | 1 | Phatr_bicluster_0239 |
| GO:0006508 | proteolysis and peptidolysis | 0.1023 | 1 | Phatr_bicluster_0239 |
| GO:0008152 | metabolism | 0.160846 | 1 | Phatr_bicluster_0239 |
| GO:0006468 | protein amino acid phosphorylation | 0.352461 | 1 | Phatr_bicluster_0239 |
| GO:0006810 | transport | 0.38354 | 1 | Phatr_bicluster_0239 |

Comments