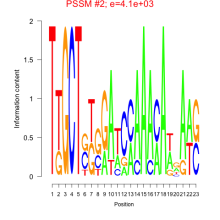
Phatr_bicluster_0257 Residual: 0.37
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0257 | 0.37 | Phaeodactylum tricornutum |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11793 flagellin-sensing 2-like protein (PLN00113)
PHATRDRAFT_16940 flag-tagged protein kinase domain ofmitogen-activated protein kinase kinase kinase (PKc_like superfamily)
PHATRDRAFT_36794 helicase domain protein (HA)
PHATRDRAFT_39238 4930451c15rik protein (LRR_RI superfamily)
PHATRDRAFT_41562 (DUF285)
PHATRDRAFT_42423 cca14_orysjcyclin-a1-4 (g2 mitotic-specific cyclin-a1-4)
PHATRDRAFT_42647 (cyclophilin superfamily)
PHATRDRAFT_44524 (PH-like superfamily)
PHATRDRAFT_45280 rna chaperone hfq
PHATRDRAFT_46346 two component regulator three y domain protein
PHATRDRAFT_46573 tpr repeat (TPR_12)
PHATRDRAFT_47709 guanylyl cyclase family member (gcy-28) (CHD)
PHATRDRAFT_47725 tropomodulin 1 (LRR_RI superfamily)
PHATRDRAFT_51953 mca1_yeastmetacaspase-1 precursor (Peptidase_C14)
PHATRDRAFT_54760 protein kinase domain containing protein (Pkinase)
PHATRDRAFT_8877 protein kinase 6-like (PKc_like superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0331273 | 1 | Phatr_bicluster_0257 |
| GO:0006468 | protein amino acid phosphorylation | 0.0424353 | 1 | Phatr_bicluster_0257 |
| GO:0000074 | regulation of cell cycle | 0.0652904 | 1 | Phatr_bicluster_0257 |
| GO:0007242 | intracellular signaling cascade | 0.0800992 | 1 | Phatr_bicluster_0257 |
| GO:0007165 | signal transduction | 0.0892501 | 1 | Phatr_bicluster_0257 |
| GO:0006508 | proteolysis and peptidolysis | 0.365994 | 1 | Phatr_bicluster_0257 |
| GO:0008152 | metabolism | 0.453158 | 1 | Phatr_bicluster_0257 |


Comments