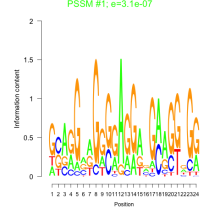
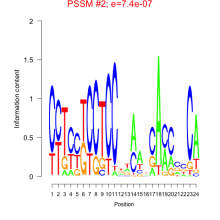
Thaps_bicluster_0096 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0096 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 36 of 36
" class="views-fluidgrid-wrapper clear-block">
10566 Na_Ca_ex
11588 hypothetical protein
12810 KAP95
13254 DEADc
16440 (RP3A) RNAP_RPB11_RPB3 superfamily
21014 hypothetical protein
21659 hypothetical protein
22311 hypothetical protein
23403 ANK
23954 hypothetical protein
24110 hypothetical protein
24680 hypothetical protein
24801 hypothetical protein
25351 hypothetical protein
25548 hypothetical protein
25701 hypothetical protein
25703 hypothetical protein
25766 hypothetical protein
32229 MogA_MoaB
32273 PCI superfamily
33476 diphth2_R
3404 hypothetical protein
34487 MgtA
34541 MgtA
35723 HepA
35933 Paf67 superfamily
37442 Ribosomal_L7Ae
38207 SecE superfamily
38472 HA2
5590 hypothetical protein
6562 hypothetical protein
7771 hypothetical protein
8940 hypothetical protein
8967 ANK
916 (Tp_CCHH2) regulator [Rayko]
9754 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015992 | proton transport | 0.00133471 | 1 | Thaps_bicluster_0096 |
| GO:0006812 | cation transport | 0.0107924 | 1 | Thaps_bicluster_0096 |
| GO:0000059 | protein-nucleus import, docking | 0.0184762 | 1 | Thaps_bicluster_0096 |
| GO:0006777 | Mo-molybdopterin cofactor biosynthesis | 0.0221325 | 1 | Thaps_bicluster_0096 |
| GO:0042254 | ribosome biogenesis and assembly | 0.025776 | 1 | Thaps_bicluster_0096 |
| GO:0006605 | protein targeting | 0.025776 | 1 | Thaps_bicluster_0096 |
| GO:0044267 | cellular protein metabolism | 0.0509236 | 1 | Thaps_bicluster_0096 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0650171 | 1 | Thaps_bicluster_0096 |
| GO:0006413 | translational initiation | 0.0857873 | 1 | Thaps_bicluster_0096 |
| GO:0006350 | transcription | 0.106121 | 1 | Thaps_bicluster_0096 |

Comments