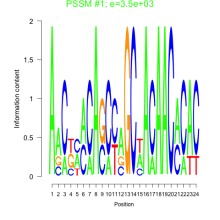
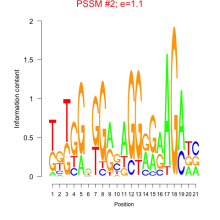
Thaps_bicluster_0102 Residual: 0.20
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0102 | 0.20 | Thalassiosira pseudonana |
Displaying 1 - 15 of 15
" class="views-fluidgrid-wrapper clear-block">
1989 FKBP_C
22194 DUF1995 superfamily
2465 hypothetical protein
264901 (Ppx1) PLN02576
2846 FDH_GDH_like superfamily
3327 hypothetical protein
33566 (TPP1) sigpep_I_bact
3506 DUF1995 superfamily
4531 PKc_like superfamily
4718 SDR_a5
5077 PLN00015
5632 hypothetical protein
6139 Chloroa_b-bind superfamily
7442 hypothetical protein
8063 SDR_c
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0102 |
| GO:0006564 | L-serine biosynthesis | 0.0166355 | 1 | Thaps_bicluster_0102 |
| GO:0008152 | metabolism | 0.171652 | 1 | Thaps_bicluster_0102 |
| GO:0009765 | photosynthesis light harvesting | 0.072069 | 1 | Thaps_bicluster_0102 |
| GO:0006779 | porphyrin biosynthesis | 0.0202987 | 1 | Thaps_bicluster_0102 |
| GO:0006468 | protein amino acid phosphorylation | 0.333459 | 1 | Thaps_bicluster_0102 |
| GO:0006457 | protein folding | 0.217609 | 1 | Thaps_bicluster_0102 |
| GO:0006508 | proteolysis and peptidolysis | 0.424754 | 1 | Thaps_bicluster_0102 |

Comments