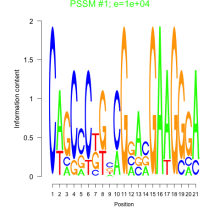
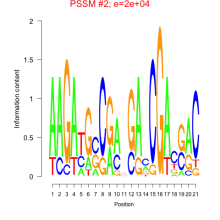
Thaps_bicluster_0105 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0105 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
10059 hypothetical protein
11479 hypothetical protein
1237 Seipin superfamily
1644 Pkinase
1871 hypothetical protein
21083 hypothetical protein
21713 UbiH
2241 Methyltransf_21
22473 aliphatic_amidase
22520 RecN
23514 hypothetical protein
24184 ArfGap
25046 hypothetical protein
26146 14-3-3
262719 CHD
2679 hypothetical protein
268472 Sugar_tr
3206 CAP10 superfamily
3450 hypothetical protein
35423 (CEN1) PTZ00183
5212 (Tp_CCCH26) regulator [Rayko]
5337 hypothetical protein
5360 hypothetical protein
6062 hypothetical protein
6838 DUF3754
8348 Peptidase_C97
940 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0001539 | ciliary or flagellar motility | 0.0123506 | 1 | Thaps_bicluster_0105 |
| GO:0006364 | rRNA processing | 0.0245587 | 1 | Thaps_bicluster_0105 |
| GO:0043087 | regulation of GTPase activity | 0.0306099 | 1 | Thaps_bicluster_0105 |
| GO:0006807 | nitrogen compound metabolism | 0.0336223 | 1 | Thaps_bicluster_0105 |
| GO:0008643 | carbohydrate transport | 0.0426071 | 1 | Thaps_bicluster_0105 |
| GO:0016192 | vesicle-mediated transport | 0.0485536 | 1 | Thaps_bicluster_0105 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.066187 | 1 | Thaps_bicluster_0105 |
| GO:0006725 | aromatic compound metabolism | 0.066187 | 1 | Thaps_bicluster_0105 |
| GO:0007242 | intracellular signaling cascade | 0.122787 | 1 | Thaps_bicluster_0105 |
| GO:0006412 | protein biosynthesis | 0.408205 | 1 | Thaps_bicluster_0105 |

Comments