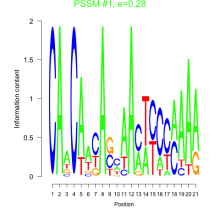
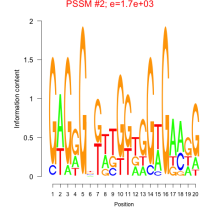
Thaps_bicluster_0118 Residual: 0.28
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0118 | 0.28 | Thalassiosira pseudonana |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
10277 hypothetical protein
1091 hypothetical protein
1219 PAP_fibrillin superfamily
1450 hypothetical protein
3166 hypothetical protein
3299 hypothetical protein
3300 hypothetical protein
33231 PLN02779
36091 DUF2499
36203 (SMT1) Sterol_MT_C superfamily
5063 hypothetical protein
5188 pyrophosphatase
5367 hypothetical protein
5785 hypothetical protein
6155 hypothetical protein
6431 hypothetical protein
6736 hypothetical protein
6905 hypothetical protein
719 PLN02433
7541 hypothetical protein
8426 hypothetical protein
8670 hypothetical protein
976 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006796 | phosphate metabolic process | 0.00124044 | 1 | Thaps_bicluster_0118 |
| GO:0006779 | porphyrin biosynthesis | 0.0135745 | 1 | Thaps_bicluster_0118 |
| GO:0019538 | protein metabolism | 0.0557488 | 1 | Thaps_bicluster_0118 |
| GO:0008152 | metabolism | 0.0844211 | 1 | Thaps_bicluster_0118 |
| GO:0006810 | transport | 0.207702 | 1 | Thaps_bicluster_0118 |

Comments