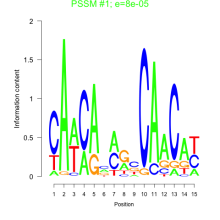
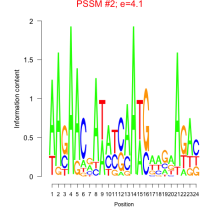
Thaps_bicluster_0130 Residual: 0.40
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0130 | 0.40 | Thalassiosira pseudonana |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
11739 hypothetical protein
12662 COG2907
1512 hypothetical protein
1626 Acid_PPase superfamily
21194 hypothetical protein
2262 SNc superfamily
23698 DUF4336 superfamily
2441 Thymidylate_kin
25513 Mnd1
261525 CypX
261582 PKCI_related
26436 Dcp
2690 WD40 superfamily
269110 hypothetical protein
30851 (CLPC2) clpA
32206 Fer4_15
38285 (PMSR1) MsrA
5288 DUF393 superfamily
5635 hypothetical protein
6122 hypothetical protein
6138 hypothetical protein
6176 hypothetical protein
7716 (Tp_CCCH27_RING95) regulator [Rayko]
8065 hypothetical protein
9483 CoAse
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006233 | dTDP biosynthesis | 0.00248088 | 1 | Thaps_bicluster_0130 |
| GO:0006235 | dTTP biosynthesis | 0.00248088 | 1 | Thaps_bicluster_0130 |
| GO:0008610 | lipid biosynthesis | 0.00742571 | 1 | Thaps_bicluster_0130 |
| GO:0006118 | electron transport | 0.024985 | 1 | Thaps_bicluster_0130 |
| GO:0006508 | proteolysis and peptidolysis | 0.0307611 | 1 | Thaps_bicluster_0130 |
| GO:0006464 | protein modification | 0.137086 | 1 | Thaps_bicluster_0130 |
| GO:0016567 | protein ubiquitination | 0.179494 | 1 | Thaps_bicluster_0130 |
| GO:0008152 | metabolism | 0.652155 | 1 | Thaps_bicluster_0130 |

Comments