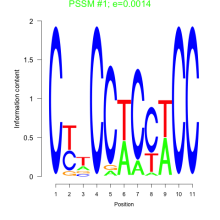
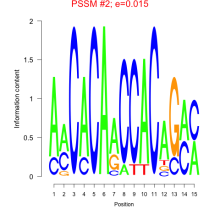
Thaps_bicluster_0165 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0165 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
12061 Lactamase_B superfamily
21163 hypothetical protein
21398 hypothetical protein
21491 hypothetical protein
22170 PCI
23539 Oligomerization
262914 (HSP90_2) HSP90 superfamily
264306 UT superfamily
268451 SrmB
268500 PRK05218
268629 PseudoU_synth_RluCD_like
31438 HEAT_2
32788 Terminase_GpA superfamily
32816 DUF155 superfamily
34221 DEXDc superfamily
34735 DRE_TIM_metallolyase superfamily
36053 Pkinase
36321 hypothetical protein
37881 DUF3172 superfamily
5667 hypothetical protein
6285 (HSP90_1) PTZ00272
7742 KTI12
9831 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006457 | protein folding | 0.0100167 | 1 | Thaps_bicluster_0165 |
| GO:0008643 | carbohydrate transport | 0.0172498 | 1 | Thaps_bicluster_0165 |
| GO:0006396 | RNA processing | 0.0698579 | 1 | Thaps_bicluster_0165 |
| GO:0006468 | protein amino acid phosphorylation | 0.236888 | 1 | Thaps_bicluster_0165 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.325408 | 1 | Thaps_bicluster_0165 |

Comments