Thaps_bicluster_0172 Residual: 0.30
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0172 | 0.30 | Thalassiosira pseudonana |
Displaying 1 - 16 of 16
" class="views-fluidgrid-wrapper clear-block">
11530 DUF1365 superfamily
15464 PCI
22248 PLN00162
24932 Thioredoxin_like superfamily
260799 Peptidase_M76
261126 DadA
34158 DUF336 superfamily
3621 hypothetical protein
36459 PLN02584
38449 hypothetical protein
4357 hypothetical protein
4981 Rhomboid superfamily
6472 hypothetical protein
8122 hypothetical protein
8333 hypothetical protein
9065 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006888 | ER to Golgi transport | 0.00618938 | 1 | Thaps_bicluster_0172 |
| GO:0006886 | intracellular protein transport | 0.0546093 | 1 | Thaps_bicluster_0172 |
| GO:0006118 | electron transport | 0.24643 | 1 | Thaps_bicluster_0172 |
| GO:0006508 | proteolysis and peptidolysis | 0.264399 | 1 | Thaps_bicluster_0172 |
| GO:0008152 | metabolism | 0.355748 | 1 | Thaps_bicluster_0172 |

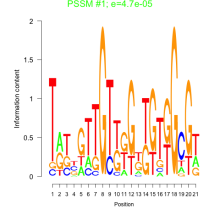

Comments