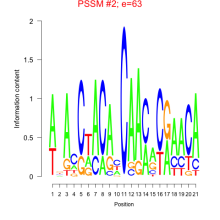
Thaps_bicluster_0198 Residual: 0.40
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0198 | 0.40 | Thalassiosira pseudonana |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
10671 hypothetical protein
15638 SET
16210 hypothetical protein
16265 S_TKc
1913 AAA
20771 hypothetical protein
20777 (Tp_TAZ1) regulator [Rayko]
21967 CCT
22122 JmjC
22324 hypothetical protein
22379 hypothetical protein
22531 hypothetical protein
22699 hypothetical protein
22709 hypothetical protein
22790 hypothetical protein
22889 HMGB-UBF_HMG-box
23345 hypothetical protein
2375 hypothetical protein
24412 hypothetical protein
24507 (Tp_bZIP5_PAS) regulator [Rayko]
270332 hypothetical protein
30866 hypothetical protein
3101 hypothetical protein
32485 PLN02681
3375 (Tp_Myb2R2) regulator [Rayko]
3447 hypothetical protein
35740 fabG
5509 Pkinase
6295 hypothetical protein
6935 (Tp_HSF_1.3h) HSF_DNA-bind superfamily
7420 hypothetical protein
7451 PAS_9
7466 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006562 | proline catabolism | 0.00186066 | 1 | Thaps_bicluster_0198 |
| GO:0006537 | glutamate biosynthesis | 0.00742418 | 1 | Thaps_bicluster_0198 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0159629 | 1 | Thaps_bicluster_0198 |
| GO:0006468 | protein amino acid phosphorylation | 0.0566487 | 1 | Thaps_bicluster_0198 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0198 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_bicluster_0198 |

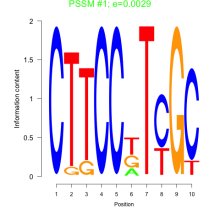
Comments