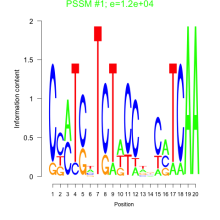
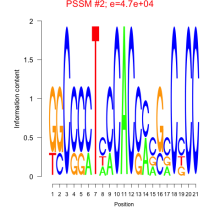
Thaps_bicluster_0213 Residual: 0.25
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0213 | 0.25 | Thalassiosira pseudonana |
Displaying 1 - 18 of 18
" class="views-fluidgrid-wrapper clear-block">
11354 hypothetical protein
12044 hypothetical protein
15791 STKc_MAPKKK
1908 hypothetical protein
20986 WcaG
2132 hypothetical protein
23028 KISc_KIF2_like
23522 DUF1032 superfamily
24403 hypothetical protein
2442 hypothetical protein
25304 hypothetical protein
268221 (Tp_Myb1R10) MYB DNA binding protein/ transcription factor-like protein
2910 Motor_domain superfamily
33955 Pkinase
37398 Motor_domain superfamily
4742 PIN_SF superfamily
5662 hypothetical protein
7967 2A0106
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007018 | microtubule-based movement | 0.000102782 | 0.281931 | Thaps_bicluster_0213 |
| GO:0006468 | protein amino acid phosphorylation | 0.0106857 | 1 | Thaps_bicluster_0213 |
| GO:0006281 | DNA repair | 0.0998737 | 1 | Thaps_bicluster_0213 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.151344 | 1 | Thaps_bicluster_0213 |
| GO:0016567 | protein ubiquitination | 0.165839 | 1 | Thaps_bicluster_0213 |
| GO:0006810 | transport | 0.347574 | 1 | Thaps_bicluster_0213 |

Comments