Thaps_bicluster_0283 Residual: 0.35
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0283 | 0.35 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
11698 hypothetical protein
12100 hypothetical protein
12103 hypothetical protein
21904 hypothetical protein
23427 hypothetical protein
23620 hypothetical protein
2378 hypothetical protein
25166 hypothetical protein
25171 DUF1517 superfamily
261641 NOX_Duox_like_FAD_NADP
263883 hypothetical protein
264384 hypothetical protein
264776 (CLPB2) chaperone_ClpB
264807 ATP-synt_C superfamily
264811 DUF389
268242 hypothetical protein
269653 hypothetical protein
269876 hypothetical protein
28025 rSAM_SeCys
30178 PLN03050
33316 (DGD1) UGD_SDR_e
34094 CdCA1
3898 hypothetical protein
40630 SPFH_like_u4
42992 (AOX2) AOX
7349 hypothetical protein
8903 DUF563
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007585 | respiratory gaseous exchange | 0.00247959 | 1 | Thaps_bicluster_0283 |
| GO:0009225 | nucleotide-sugar metabolism | 0.0111179 | 1 | Thaps_bicluster_0283 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0282075 | 1 | Thaps_bicluster_0283 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0557488 | 1 | Thaps_bicluster_0283 |
| GO:0019538 | protein metabolism | 0.0557488 | 1 | Thaps_bicluster_0283 |
| GO:0006118 | electron transport | 0.287914 | 1 | Thaps_bicluster_0283 |


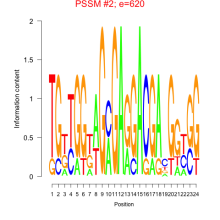
Comments