Regulatory Modules
Displaying 61 - 70 of 170Desulfovibrio vulgaris Hildenborough
| Title | Motif 1 | Motif 2 | Residual | Bicluster Genes | Selected GO Terms |
|---|---|---|---|---|---|
| bicluster_0282 |
0.18 |
500 |
0.45 | DVU0551, DVU0789, DVU1291, DVU1583, DVU1620, DVU1673, DVU2890, DVU0734, DVU1082, DVU1292, DVU1579, DVU1582, DVU1584, DVU1587, DVU1663, DVU1672, DVU1818, DVU1828 | cell morphogenesis, anatomical structure morphogenesis, developmental process, cellular component morphogenesis, single-organism developmental process |
| bicluster_0191 |
0.0000000000000046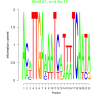 |
0.31 |
0.45 | DVU3122, DVU0273, DVU0303, DVU0304, DVU0762, DVU0763, DVU2377, DVU2378, DVU2379, DVU2380, DVU2383, DVU2388, DVU2389, DVU2390, DVU2571, DVU2572, DVU2573, DVU2574, DVU2680, DVU2681, DVU3331, DVU3332, DVU3333 | ion binding, metal ion binding, cation binding, transition metal ion binding, binding |
| bicluster_0090 |
0.2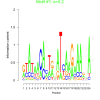 |
1400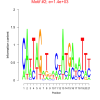 |
0.45 | DVU0223, DVU0526, DVU0727, DVU0728, DVU0756, DVU1151, DVU1492, DVU1628, DVU1630, DVU1710, DVU1730, DVU1855, DVU2309, DVU2423, DVU2845, DVU2846, DVUA0089, DVU1379, DVU1629, DVU1729, DVU2614 | |
| bicluster_0004 |
3.1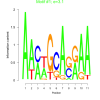 |
2600 |
0.45 | DVU0483, DVU0595, DVU0662, DVU0663, DVU0664, DVU0725, DVU0812, DVU1013, DVU1212, DVU1629, DVU1839, DVU1874, DVU2324, DVU2439, DVU2467, DVU2468, DVU2469, DVU2974, DVU3283, DVU3316 | sulfur amino acid metabolic process, sulfur amino acid biosynthetic process, serine family amino acid metabolic proce..., cellular amino acid metabolic process, O-acyltransferase activity |
| bicluster_0008 |
0.00000000018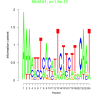 |
0.00000000027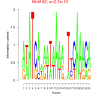 |
0.45 | DVU0252, DVU0268, DVU0472, DVU0473, DVU0553, DVU0630, DVU0916, DVU0939, DVU1015, DVU1155, DVU1285, DVU1637, DVU1638, DVU1639, DVU1646, DVU2145, DVU2146, DVU2725, DVU2726, DVU3080 | O-acyltransferase activity, acetyltransferase activity |
| bicluster_0197 |
11000 |
13000 |
0.45 | DVU1724, DVU2706, DVU3142, DVUA0012, DVUA0034, DVUA0038, DVUA0042, DVUA0054, DVU0621, DVU2692, DVU2881, DVUA0031, DVUA0032, DVUA0035, DVUA0093, DVUA0102, DVUA0128, DVUA0136 | inorganic anion transmembrane transporte..., oxidoreductase activity, acting on iron-..., oxidoreductase activity, anion transmembrane transporter activity |
| bicluster_0299 |
4500 |
6800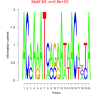 |
0.44 | DVU0442, DVU1970, DVU0185, DVU0443, DVU0445, DVU1105, DVU1414, DVU1503, DVU1721, DVU1967, DVU1968, DVU1969, DVU2168, DVU2171, DVU2173, DVU2187 | [protein-PII] uridylyltransferase activi..., uridylyltransferase activity, exonuclease activity, nucleotidyltransferase activity |
| bicluster_0339 |
0.000006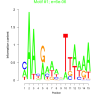 |
0.19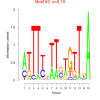 |
0.44 | DVU0371, DVU0554, DVU0555, DVU0556, DVU0558, DVU0559, DVU0561, DVU0562, DVU0923, DVU1147, DVU1166, DVU1167, DVU1277, DVU1832, DVU2010, DVU2048, DVU2175, DVU2827, DVU3089 | DNA integration, carbohydrate metabolic process |
| bicluster_0034 |
0.0000000000000014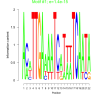 |
0.15 |
0.44 | DVU0273, DVU0303, DVU0304, DVU0762, DVU0763, DVU2377, DVU2378, DVU2383, DVU2564, DVU2571, DVU2572, DVU2573, DVU2574, DVU2680, DVU2681, DVU3124, DVU3331, DVU3332, DVU3333, DVU2381, DVU3330 | transition metal ion binding, metal ion binding, cation binding, ion binding |
| bicluster_0203 |
1100 |
4100 |
0.44 | DVU2062, DVU2218, DVU2424, DVU2517, DVU1385, DVU1431, DVU1836, DVU2295, DVU2906, DVU3132, DVU3142 | response to stimulus, cellular response to stimulus, DNA repair, cellular response to DNA damage stimulus, catalytic complex |

