Cre01.g051800.t1.1 Pyridoxal phosphate (PLP)-dependent transferases superfamily protein
Details for Cre01.g051800.t1.1 - Pyridoxal phosphate (PLP)-dependent transferases superfamily protein
GO Terms: GO:0030170, GO:0009058
Go to Gene Page: Cre01.g051800.t1.1
Phytozome Genomic Region

|
Cre02.g086150.t1.2
Details for Cre02.g086150.t1.2 -
GO Terms: NA
Go to Gene Page: Cre02.g086150.t1.2
Phytozome Genomic Region

|
Cre02.g095294.t1.2 Zinc finger (C3HC4-type RING finger) family protein
Details for Cre02.g095294.t1.2 - Zinc finger (C3HC4-type RING finger) family protein
GO Terms: NA
Go to Gene Page: Cre02.g095294.t1.2
Phytozome Genomic Region

|
Cre02.g145950.t1.2
Details for Cre02.g145950.t1.2 -
GO Terms: NA
Go to Gene Page: Cre02.g145950.t1.2
Phytozome Genomic Region

|
Cre03.g161578.t4.1
Details for Cre03.g161578.t4.1 -
GO Terms: NA
Go to Gene Page: Cre03.g161578.t4.1
Phytozome Genomic Region

|
Cre03.g194700.t1.1 alpha-xylosidase 1
|
|
Cre05.g240000.t1.2 lipid phosphate phosphatase 3
|
Cre06.g260650.t1.1 Protein of unknown function (DUF288)
Details for Cre06.g260650.t1.1 - Protein of unknown function (DUF288)
GO Terms: NA
Go to Gene Page: Cre06.g260650.t1.1
Phytozome Genomic Region

|
Cre06.g310900.t1.2
Details for Cre06.g310900.t1.2 -
GO Terms: NA
Go to Gene Page: Cre06.g310900.t1.2
Phytozome Genomic Region

|
Cre06.g311700.t1.2
Details for Cre06.g311700.t1.2 -
GO Terms: NA
Go to Gene Page: Cre06.g311700.t1.2
Phytozome Genomic Region

|
|
Cre09.g399050.t1.1 SGNH hydrolase-type esterase superfamily protein
|
Cre09.g416309.t1.1
Details for Cre09.g416309.t1.1 -
GO Terms: NA
Go to Gene Page: Cre09.g416309.t1.1
Phytozome Genomic Region

|
Cre12.g493050.t1.2
Details for Cre12.g493050.t1.2 -
GO Terms: NA
Go to Gene Page: Cre12.g493050.t1.2
Phytozome Genomic Region

|
Cre12.g529450.t2.1 Dynamin related protein 5A
|
Cre12.g536050.t1.2 ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein
Details for Cre12.g536050.t1.2 - ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein
GO Terms: GO:0000166, GO:0046872
Go to Gene Page: Cre12.g536050.t1.2
Phytozome Genomic Region

|
Cre12.g556228.t1.1 FAD/NAD(P)-binding oxidoreductase family protein
|
Cre13.g576466.t1.1
Details for Cre13.g576466.t1.1 -
GO Terms: NA
Go to Gene Page: Cre13.g576466.t1.1
Phytozome Genomic Region

|
Cre14.g624800.t1.2 ARRESTIN DOMAIN CONTAINING PROTEIN
Details for Cre14.g624800.t1.2 - ARRESTIN DOMAIN CONTAINING PROTEIN
GO Terms: NA
Go to Gene Page: Cre14.g624800.t1.2
Phytozome Genomic Region

|
Cre14.g625850.t1.1
Details for Cre14.g625850.t1.1 -
GO Terms: NA
Go to Gene Page: Cre14.g625850.t1.1
Phytozome Genomic Region

|
Cre16.g681350.t1.1
Details for Cre16.g681350.t1.1 -
GO Terms: NA
Go to Gene Page: Cre16.g681350.t1.1
Phytozome Genomic Region

|
Cre17.g699500.t1.2 tubulin-tyrosine ligases;tubulin-tyrosine ligases
Details for Cre17.g699500.t1.2 - tubulin-tyrosine ligases;tubulin-tyrosine ligases
GO Terms: GO:0006464
Go to Gene Page: Cre17.g699500.t1.2
Phytozome Genomic Region

|
Cre17.g701350.t3.1 Polynucleotidyl transferase, ribonuclease H-like superfamily protein
Details for Cre17.g701350.t3.1 - Polynucleotidyl transferase, ribonuclease H-like superfamily protein
GO Terms: NA
Go to Gene Page: Cre17.g701350.t3.1
Phytozome Genomic Region

|
Cre17.g702300.t1.1
Details for Cre17.g702300.t1.1 -
GO Terms: NA
Go to Gene Page: Cre17.g702300.t1.1
Phytozome Genomic Region

|
Cre17.g708400.t1.2
|
Cre36.g759747.t1.1
Details for Cre36.g759747.t1.1 -
GO Terms: NA
Go to Gene Page: Cre36.g759747.t1.1
Phytozome Genomic Region

|
|

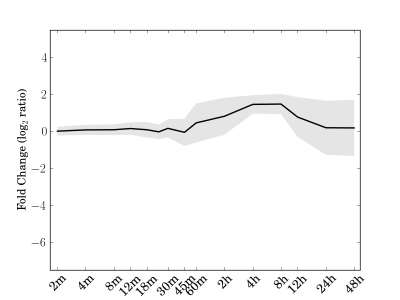
Comments