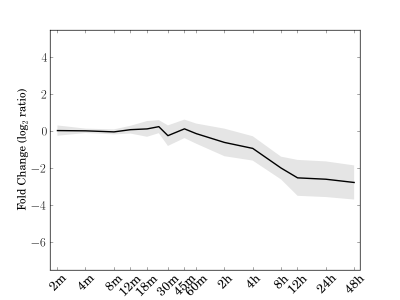
Module 33 Residual: 0.10
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0033 | v02 | 0.10 | -15.89 |
Displaying 1 - 23 of 23
| Cre01.g010900.t1.2 glyceraldehyde-3-phosphate dehydrogenase B subunit | Cre01.g026900.t1.2 FAD/NAD(P)-binding oxidoreductase family protein |
| Cre02.g144007.t1.1 | Cre02.g144500.t1.1 KH domain-containing protein |
| Cre06.g265500.t1.2 Histone superfamily protein | Cre07.g328075.t1.1 |
| Cre08.g366050.t1.2 nudix hydrolase homolog 3 | Cre10.g424450.t1.2 translocase of the outer mitochondrial membrane 40 |
| Cre10.g431850.t1.2 Phosphoglycerate mutase family protein | Cre10.g440050.t1.2 chloroplast stem-loop binding protein of 41 kDa |
| Cre11.g467569.t1.1 ATP synthase delta-subunit gene | Cre11.g467689.t1.1 photosynthetic electron transfer C |
| Cre12.g510050.t1.2 dicarboxylate diiron protein, putative (Crd1) | Cre12.g529800.t1.1 RNI-like superfamily protein |
| Cre12.g544050.t1.1 poly(A) binding protein 8 | Cre13.g562150.t1.2 GTP-binding protein-related |
| Cre13.g573250.t1.2 sulfurtransferase protein 16 | Cre13.g581450.t1.2 26S proteasome, regulatory subunit Rpn7;Proteasome component (PCI) domain |
| Cre13.g588100.t1.2 rotamase cyclophilin 2 | Cre14.g629800.t1.2 |
| Cre16.g650800.t1.1 translocase of the inner mitochondrial membrane 13 | Cre16.g677450.t1.2 Galactose mutarotase-like superfamily protein |
| Cre16.g682900.t2.1 CLP protease proteolytic subunit 2 |
|
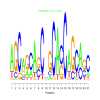
e.value: 0.000011 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: AGCagcAgCnGCAcCAGCagC |
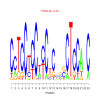
motif_0033_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 29 Motif Bicluster: Width: 23 Number of Sites: 1 Consensus: CcTCCtCCtcCtCcgcCTCCAnC |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
| up-regulates |
Cre06.g268600.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51glycine-rich protein 2B |
0.048991 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

Comments