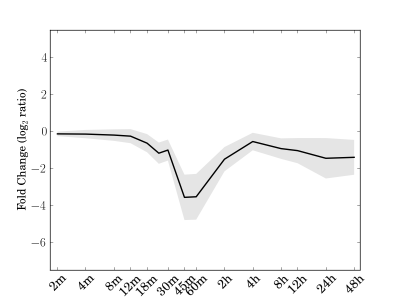
Module 71 Residual: 0.17
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0071 | v02 | 0.17 | -14.47 |
Displaying 1 - 18 of 18
| Cre01.g044400.t1.2 | Cre02.g083600.t2.1 GTP binding |
| Cre03.g172550.t1.2 arginine methyltransferase 11 | Cre06.g296700.t1.2 Hydrogenase assembly factor/biotin synthase |
| Cre06.g309450.t1.2 carboxypeptidase D, putative | Cre07.g351750.t1.2 rRNA processing protein-related |
| Cre07.g356600.t1.2 | Cre09.g394436.t1.1 Inorganic H pyrophosphatase family protein |
| Cre09.g416800.t1.2 | Cre10.g452350.t1.1 |
| Cre12.g529800.t2.1 RNI-like superfamily protein | Cre12.g541400.t1.2 |
| Cre12.g558100.t1.2 protein arginine methyltransferase 10 | Cre13.g586700.t1.1 |
| Cre16.g663800.t1.2 Nucleotide/sugar transporter family protein | Cre16.g679450.t1.2 PIN domain-like family protein |
| Cre17.g708100.t1.2 Hypoxanthine-guanine phosphoribosyltransferase | Cre17.g742000.t1.2 Predicted protein |
|
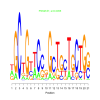
e.value: 0.0095 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: nGCtGttCcgGctgcTgCTGg |
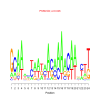
motif_0071_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.026 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: GcAAactaataAcaaAacAanctt |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

Comments