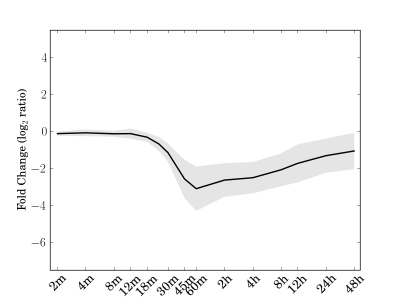
Module 84 Residual: 0.15
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0084 | v02 | 0.15 | -11.76 |
Displaying 1 - 21 of 21
| Cre01.g015600.t1.1 ILITYHIA | Cre01.g039750.t1.2 peptide deformylase 1B |
| Cre01.g047800.t1.1 CAX-interacting protein 2 | Cre02.g103850.t1.2 HISTIDINE BIOSYNTHESIS 5B |
| Cre03.g145747.t1.1 chorismate synthase, putative / 5-enolpyruvylshikimate-3-phosphate phospholyase, putative | Cre03.g156050.t1.2 ribosome recycling factor, chloroplast precursor |
| Cre03.g164700.t1.2 | Cre03.g177200.t1.2 RNA-binding (RRM/RBD/RNP motifs) family protein |
| Cre04.g218700.t1.2 Nucleic acid-binding, OB-fold-like protein | Cre06.g266250.t1.2 Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain |
| Cre06.g301650.t1.2 | Cre09.g396252.t1.1 glutathione reductase |
| Cre11.g479150.t3.1 | Cre12.g507800.t1.2 |
| Cre12.g541777.t1.1 Rubisco methyltransferase family protein | Cre13.g580000.t1.1 |
| Cre13.g580850.t1.2 ribosomal protein L22 | Cre14.g629650.t1.1 high-affinity nickel-transport family protein |
| Cre16.g676314.t1.1 translation initiation factor 3 subunit H1 | Cre16.g684150.t1.2 |
| Cre16.g688050.t1.1 eukaryotic translation initiation factor 4B1 |
|
e.value: 0.033 Motif Bicluster: Width: 14 Number of Sites: 1 Consensus: TaaTTGTtaTTgTT |
motif_0084_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 79 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: GCAgCGgCAGCagcA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0004424 | imidazoleglycerol-phosphate dehydratase ... |

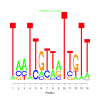
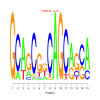
Comments