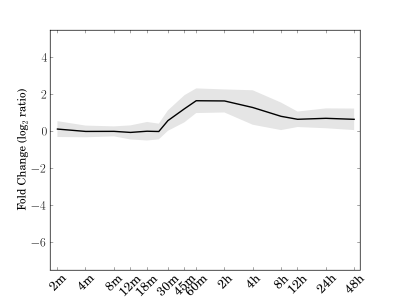
Module 101 Residual: 0.34
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0101 | v02 | 0.34 | -15.28 |
Displaying 1 - 25 of 25
| Cre03.g150200.t1.2 | Cre03.g187850.t1.2 |
| Cre03.g192550.t1.1 DNA repair-recombination protein (RAD50) | Cre03.g197850.t2.1 |
| Cre04.g217240.t1.1 | Cre06.g277650.t1.2 Bestrophin-like protein |
| Cre06.g278144.t1.1 | Cre06.g281000.t1.1 SET domain-containing protein |
| Cre06.g290800.t1.1 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Cre07.g315550.t1.1 AFG1-like ATPase family protein |
| Cre07.g320850.t1.2 beta glucosidase 42 | Cre07.g357350.t1.2 GLNB1 homolog |
| Cre08.g370200.t1.2 trithorax-like protein 2 | Cre10.g421300.t1.2 |
| Cre10.g437000.t1.2 | Cre11.g467655.t1.1 Peptidase S24/S26A/S26B/S26C family protein |
| Cre11.g476650.t1.1 limit dextrinase | Cre12.g493500.t1.2 FAD-dependent oxidoreductase family protein |
| Cre12.g499550.t1.2 | Cre12.g527350.t1.2 |
| Cre12.g541800.t1.2 Prolyl oligopeptidase family protein | Cre16.g654050.t1.1 |
| Cre16.g687117.t1.2 | Cre16.g687650.t1.1 Ankyrin repeat family protein |
| Cre16.g694850.t1.2 arginine biosynthesis protein ArgJ family |
|
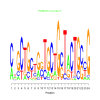
e.value: 4.4e-17 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: cnGCTGctggTGCtGcTaCTGccG |
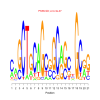
motif_0101_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.00000043 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: caGCTgCAGcgGCaGctGCgg |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0004358 | glutamate N-acetyltransferase activity |

Comments