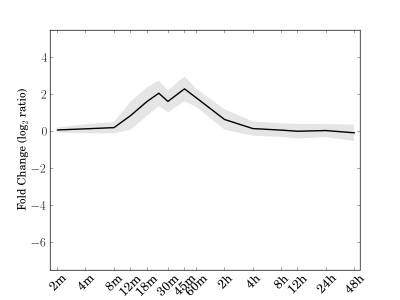
Module 138 Residual: 0.23
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0138 | v02 | 0.23 | -14.00 |
Displaying 1 - 23 of 23
| Cre01.g033150.t1.2 Domain of unknown function (DUF220) | Cre02.g089237.t1.1 proteolysis 6 |
| Cre02.g102900.t1.1 ACT-like protein tyrosine kinase family protein | Cre03.g175700.t1.2 plastid transcriptionally active 17 |
| Cre03.g193700.t1.1 alpha/beta-Hydrolases superfamily protein | Cre06.g293051.t1.1 ammonium transporter 1;1 |
| Cre07.g333150.t1.2 | Cre08.g358532.t1.1 GATA transcription factor 9 |
| Cre08.g363300.t1.2 | Cre08.g378650.t1.2 |
| Cre09.g387801.t1.1 SPla/RYanodine receptor (SPRY) domain-containing protein | Cre09.g389356.t1.1 permease, cytosine/purines, uracil, thiamine, allantoin family protein |
| Cre09.g400150.t1.1 acyl-CoA binding protein 5 | Cre12.g512950.t1.2 Chromate ion transporter |
| Cre12.g541211.t1.1 | Cre12.g548900.t1.1 Pleckstrin homology (PH) domain superfamily protein |
| Cre13.g569600.t1.2 | Cre16.g651050.t1.2 Cytochrome c |
| Cre16.g681750.t2.1 autoinhibited Ca2+ -ATPase, isoform 8 | Cre16.g683483.t1.1 DNA nucleotide excision repair protein |
| Cre17.g733678.t1.2 | Cre19.g750847.t3.1 SPFH/Band 7/PHB domain-containing membrane-associated protein family |
| Cre24.g755847.t1.1 metallo-beta-lactamase family protein |
|
e.value: 3e-37 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: ACACaCACaCACACACACACACaC |
motif_0138_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.00000000013 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: ACACACacaCACACA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0000255 | GO:0000256 | GO:0015703 | GO:0043605 | GO:0004848 | GO:0015109 | allantoin metabolic process | allantoin catabolic process | chromate transport | cellular amide catabolic process | ureidoglycolate hydrolase activity | chromate transmembrane transporter activ... |



Comments