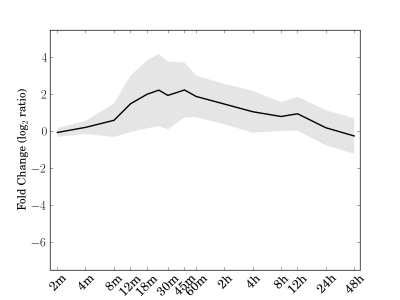
Module 176 Residual: 0.39
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0176 | v02 | 0.39 | -11.18 |
Displaying 1 - 29 of 29
| Cre01.g018850.t1.2 DNAse I-like superfamily protein | Cre01.g024500.t1.1 |
| Cre01.g044550.t1.2 | Cre02.g097800.t1.1 multidrug resistance-associated protein 2 |
| Cre02.g099750.t1.1 Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain | Cre02.g141550.t1.2 ATP-dependent protease La (LON) domain protein |
| Cre03.g144084.t1.1 RHOMBOID-like 2 | Cre03.g145267.t1.1 |
| Cre03.g180600.t1.1 | Cre03.g197750.t1.2 glutathione peroxidase 1 |
| Cre04.g223300.t1.2 Mitochondrial substrate carrier family protein | Cre05.g237050.t1.1 Protein of unknown function (DUF1230) |
| Cre05.g238311.t1.1 | Cre05.g248450.t1.2 beta carbonic anhydrase 6 |
| Cre06.g269950.t1.2 ATPase, AAA-type, CDC48 protein | Cre06.g281600.t1.2 Low-CO2-inducible protein, septin-like |
| Cre06.g288800.t1.2 Haloacid dehalogenase-like hydrolase (HAD) superfamily protein | Cre06.g289500.t1.2 |
| Cre06.g306200.t1.1 | Cre08.g359133.t1.1 |
| Cre09.g414100.t1.2 | Cre10.g466500.t1.2 Lactoylglutathione lyase / glyoxalase I family protein |
| Cre12.g505400.t1.2 | Cre12.g540250.t1.2 Selenoprotein W1 |
| Cre13.g568400.t1.2 FTSH protease 4 | Cre14.g634100.t1.1 |
| Cre16.g683931.t1.1 | Cre17.g721500.t1.2 UDP-Glycosyltransferase superfamily protein |
| Cre17.g735950.t1.2 transthyretin-like protein |
|
e.value: 0.000000000000064 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CAGCACCAgCagCAgcagCAc |
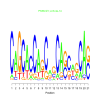
motif_0176_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.00000039 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: tGGcgGtGGcGGCaG |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

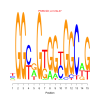
Comments