Module 189 Residual: 0.11
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0189 | v02 | 0.11 | -14.19 |
Displaying 1 - 16 of 16
| Cre01.g016514.t1.1 lipoamide dehydrogenase 1 | Cre01.g016900.t1.2 |
| Cre01.g040300.t1.2 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Cre01.g046652.t1.1 Nucleic acid-binding, OB-fold-like protein |
| Cre02.g092400.t1.1 Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain | Cre06.g250200.t1.2 methionine adenosyltransferase 3 |
| Cre06.g251900.t1.2 63 kDa inner membrane family protein | Cre06.g257601.t1.2 2-cysteine peroxiredoxin B |
| Cre06.g278195.t1.1 | Cre08.g359350.t1.2 acetyl Co-enzyme a carboxylase biotin carboxylase subunit |
| Cre08.g362900.t1.1 Photosystem II reaction center PsbP family protein | Cre11.g467770.t1.1 Phosphoglycerate kinase family protein |
| Cre12.g484000.t1.2 acetyl-CoA carboxylase carboxyl transferase subunit beta | Cre12.g498550.t1.1 magnesium-protoporphyrin IX methyltransferase |
| Cre13.g562850.t1.2 photosystem II reaction center PSB29 protein | Cre17.g701700.t2.1 Plant stearoyl-acyl-carrier-protein desaturase family protein |
|
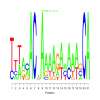
e.value: 31 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: TttacACnAaaAgaaaaagCA |
motif_0189_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 87000 Motif Bicluster: Width: 23 Number of Sites: 1 Consensus: AGCCCCCAAcCCCCCcAAAaCgT |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0044802 | GO:0061024 | single-organism membrane organization | membrane organization |



Comments