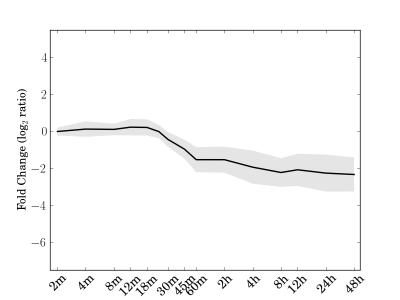
Module 201 Residual: 0.18
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0201 | v02 | 0.18 | -13.16 |
Displaying 1 - 21 of 21
| Cre01.g029300.t1.2 triosephosphate isomerase | Cre01.g052250.t1.2 thioredoxin X |
| Cre02.g074900.t1.1 Calcium-dependent lipid-binding (CaLB domain) family protein | Cre02.g098450.t1.2 eukaryotic translation initiation factor 2 gamma subunit |
| Cre02.g105650.t1.2 low psii accumulation2 | Cre03.g151200.t1.2 Predicted protein |
| Cre03.g182500.t1.2 SRP72 RNA-binding domain | Cre03.g183100.t1.2 Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein |
| Cre03.g205900.t1.2 S-adenosylmethionine decarboxylase | Cre06.g278148.t1.1 glyoxylate reductase 2 |
| Cre06.g280750.t1.1 | Cre06.g284650.t1.2 Dual-specificity protein phosphatase |
| Cre06.g308533.t1.1 | Cre08.g358540.t1.1 NAD(P)-linked oxidoreductase superfamily protein |
| Cre08.g371650.t1.2 Bacterial sec-independent translocation protein mttA/Hcf106 | Cre10.g436500.t1.2 Survival protein SurE-like phosphatase/nucleotidase |
| Cre10.g458850.t1.2 | Cre11.g467723.t2.1 3-ketoacyl-acyl carrier protein synthase I |
| Cre11.g479900.t1.2 | Cre12.g522600.t1.2 cytochrome c-2 |
| Cre13.g604000.t1.1 |
|
e.value: 0.25 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: TGntGgtGgTGGTgG |
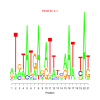
motif_0201_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:17e.value: 1 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: aTntATgAgaTttnaAtntAt |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

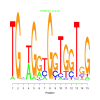
Comments