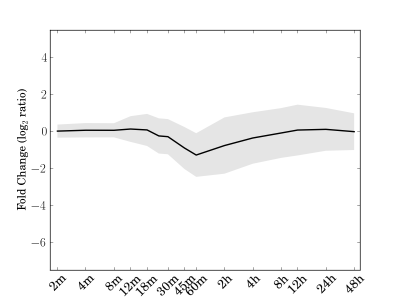
Module 19 Residual: 0.27
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0019 | v02 | 0.27 | -11.43 |
Displaying 1 - 29 of 29
| Cre01.g023900.t1.1 | Cre01.g029850.t1.2 Protein kinase family protein |
| Cre01.g049826.t1.1 | Cre02.g076600.t1.2 Peptidyl-tRNA hydrolase family protein |
| Cre02.g076950.t1.1 subtilase 4.13 | Cre02.g083600.t2.1 GTP binding |
| Cre02.g091550.t1.2 ssDNA-binding transcriptional regulator | Cre03.g154400.t1.2 |
| Cre03.g166750.t1.2 Protein kinase superfamily protein | Cre05.g238250.t1.2 Basic-leucine zipper (bZIP) transcription factor family protein |
| Cre05.g240000.t1.2 lipid phosphate phosphatase 3 | Cre06.g297400.t1.1 Oxidoreductase, zinc-binding dehydrogenase family protein |
| Cre08.g358350.t1.1 | Cre09.g389250.t1.2 LSD1-like 3 |
| Cre09.g396846.t1.1 Ypt/Rab-GAP domain of gyp1p superfamily protein | Cre09.g405750.t1.1 carbonic anhydrase 1 |
| Cre09.g407200.t1.2 FAD/NAD(P)-binding oxidoreductase family protein | Cre09.g407850.t1.2 |
| Cre12.g495954.t1.1 | Cre12.g536050.t1.2 ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein |
| Cre12.g558000.t1.1 Translation elongation factor EF1B/ribosomal protein S6 family protein | Cre13.g586650.t1.1 FixL-like PAS domain protein |
| Cre14.g624476.t1.1 | Cre16.g656100.t1.1 |
| Cre16.g662702.t1.1 RNA-binding (RRM/RBD/RNP motifs) family protein | Cre16.g685100.t1.1 Cobalamin biosynthesis CobW-like protein |
| Cre17.g699500.t1.2 tubulin-tyrosine ligases;tubulin-tyrosine ligases | Cre17.g712000.t1.1 RS-containing zinc finger protein 21 |
| Cre17.g716500.t2.1 |
|
e.value: 0.029 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: tttgcAAAtgCTtatAtgnnt |
motif_0019_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 7e-35 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: TGcTggtGCTGnTgCtGCTGc |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |


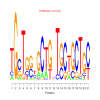
Comments