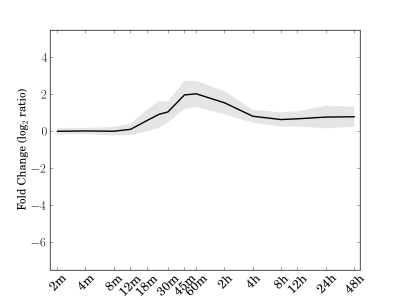
Module 63 Residual: 0.22
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0063 | v02 | 0.22 | -15.91 |
Displaying 1 - 27 of 27
| Cre01.g007150.t1.1 SPla/RYanodine receptor (SPRY) domain-containing protein | Cre02.g113100.t1.2 Uncharacterised protein family (UPF0041) |
| Cre03.g183650.t1.1 PLC-like phosphodiesterases superfamily protein | Cre05.g243550.t1.2 |
| Cre06.g278144.t1.1 | Cre06.g280300.t1.1 ubiquitin protein ligase 6 |
| Cre06.g282000.t2.1 starch synthase 3 | Cre07.g314000.t1.1 |
| Cre07.g334700.t1.2 | Cre08.g358522.t1.1 xylem cysteine peptidase 1 |
| Cre09.g398350.t1.2 U-box domain-containing protein | Cre10.g433400.t1.2 |
| Cre10.g456100.t1.2 flavodoxin-like quinone reductase 1 | Cre10.g464900.t1.1 |
| Cre11.g467609.t1.1 ubiquitin-protein ligase 2 | Cre11.g467655.t1.1 Peptidase S24/S26A/S26B/S26C family protein |
| Cre12.g541800.t1.2 Prolyl oligopeptidase family protein | Cre12.g554850.t1.2 thioredoxin H-type 1 |
| Cre13.g575850.t1.2 NAD(P)-binding Rossmann-fold superfamily protein | Cre13.g589700.t1.2 vacuolar protein sorting-associated protein 20.2 |
| Cre13.g602901.t1.1 cofactor of nitrate reductase and xanthine dehydrogenase 2 | Cre16.g647950.t1.1 |
| Cre16.g662250.t1.1 | Cre16.g673281.t1.1 phytanoyl-CoA dioxygenase (PhyH) family protein |
| Cre16.g676850.t1.2 P-loop containing nucleoside triphosphate hydrolases superfamily protein | Cre16.g687117.t1.2 |
| Cre16.g694850.t1.2 arginine biosynthesis protein ArgJ family |
|
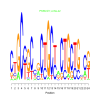
e.value: 6e-22 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: CtGCcGCcgCtGCtaCTGCtGccg |
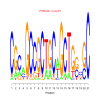
motif_0063_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.000000002 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CGcaGCCGCTGCaGCTGgcGC |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0004358 | glutamate N-acetyltransferase activity |

Comments