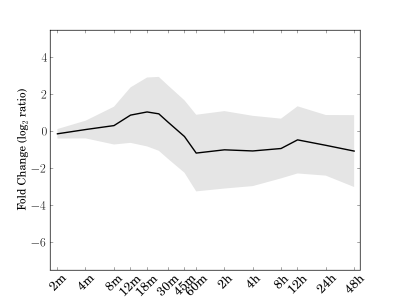
Module 121 Residual: 0.29
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0121 | v02 | 0.29 | -13.71 |
Displaying 1 - 23 of 23
| Cre02.g080800.t1.2 | Cre02.g099400.t1.1 exostosin family protein |
| Cre02.g107950.t1.2 | Cre03.g159500.t2.1 Pyridoxal-dependent decarboxylase family protein |
| Cre03.g167051.t1.2 pyridoxine biosynthesis 2 | Cre03.g175700.t1.2 plastid transcriptionally active 17 |
| Cre04.g225050.t1.1 | Cre06.g258700.t1.1 methylcrotonyl-CoA carboxylase alpha chain, mitochondrial / 3-methylcrotonyl-CoA carboxylase 1 (MCCA) |
| Cre07.g340850.t1.2 cytochrome P450, family 711, subfamily A, polypeptide 1 | Cre08.g367400.t1.1 chlorophyll A/B binding protein 1 |
| Cre08.g367500.t1.1 chlorophyll A/B binding protein 1 | Cre09.g389393.t1.1 |
| Cre09.g394750.t1.2 ribosomal protein S1 | Cre10.g440450.t1.2 photosystem II reaction center PSB28 protein |
| Cre12.g501600.t1.2 Basic-leucine zipper (bZIP) transcription factor family protein | Cre12.g533351.t1.1 heat shock protein 101 |
| Cre14.g611450.t1.1 Plastid-lipid associated protein PAP / fibrillin family protein | Cre16.g648300.t1.2 phosphate transporter 4;5 |
| Cre16.g672385.t1.1 histidinol phosphate aminotransferase 1 | Cre16.g677750.t1.2 |
| Cre16.g683550.t1.2 P450 reductase 1 | Cre17.g697406.t1.2 |
| Cre19.g751047.t1.1 |
|
e.value: 0.00000032 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: cAgnTatGTAtgTGTgggTat |
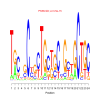
motif_0121_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.0000000000000035 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: TgCngCcgcTGctgCcGctGCTGC |
Displaying 1 - 2 of 2
| Interaction | Weight | |
|---|---|---|
| up-regulates |
Cre12.g501600.t1.2Submitted by admin on Tue, 05/19/2015 - 15:51Basic-leucine zipper (bZIP) transcription factor family protein |
0.030496 |
| up-regulates |
Cre17.g746547.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51Basic-leucine zipper (bZIP) transcription factor family protein |
0.026623 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |

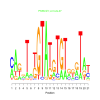
Comments