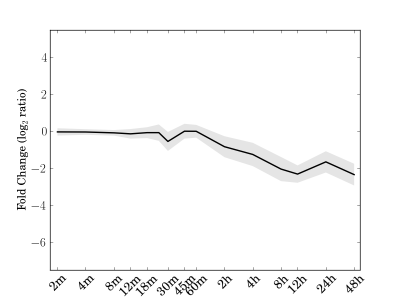
Module 149 Residual: 0.23
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0149 | v02 | 0.23 | -15.92 |
Displaying 1 - 13 of 13
| Cre01.g026900.t1.2 FAD/NAD(P)-binding oxidoreductase family protein | Cre03.g144667.t1.1 ClpS-like protein |
| Cre03.g174750.t1.2 Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein | Cre03.g183750.t1.1 |
| Cre03.g207000.t1.2 abscisic acid (aba)-deficient 4 | Cre06.g278236.t1.1 S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| Cre06.g290300.t1.2 | Cre07.g335700.t1.2 |
| Cre08.g360550.t1.1 ERD4-related membrane protein | Cre08.g368450.t2.1 ribophorin II (RPN2) family protein |
| Cre12.g519000.t1.1 phosphoribosylanthranilate isomerase 1 | Cre13.g564050.t1.1 Mog1/PsbP/DUF1795-like photosystem II reaction center PsbP family protein |
| Cre16.g669650.t1.2 endoribonuclease L-PSP family protein |
|
e.value: 0.28 Motif Bicluster: Width: 20 Number of Sites: 1 Consensus: AcAAtTATgcaAgtatGGaa |
motif_0149_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.000025 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: AgcAgcaGgnggGgCaGGaGCtGg |
Displaying 1 - 3 of 3
| Interaction | Weight | |
|---|---|---|
| up-regulates |
Cre09.g386753.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51 |
0.06633 |
| down-regulates |
Cre12.g523000.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51zinc finger protein 1 |
0.054967 |
| down-regulates |
Cre08.g375400.t1.1Submitted by admin on Tue, 05/19/2015 - 15:51KNOTTED-like homeobox of Arabidopsis thaliana 7 |
0.064381 |
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| Not available |


Comments