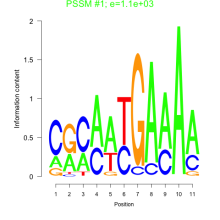
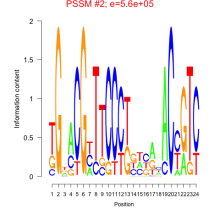
Phatr_bicluster_0093 Residual: 0.28
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0093 | 0.28 | Phaeodactylum tricornutum |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10779 branched-chain amino acid aminotransferase (PRK13357)
PHATRDRAFT_11189 translation elongation factor g (PRK00007)
PHATRDRAFT_15211 glycine-rich protein 2 (CSP_CDS)
PHATRDRAFT_20809 at5g66120 k2a18_20 (DHQS)
PHATRDRAFT_22909 ll-diaminopimelate aminotransferase (COG0436)
PHATRDRAFT_29885 vitamin b6 biosynthesis protein (pdxS)
PHATRDRAFT_32916 had superfamily (subfamily ia)tigr01548 (COG0637)
PHATRDRAFT_41511 translation elongation factor p (PRK00529)
PHATRDRAFT_43464 (DRIM superfamily)
PHATRDRAFT_45243 amine oxidase (COG1233)
PHATRDRAFT_45701 1 aminotransferase (AAT_I superfamily)
PHATRDRAFT_45964 atpcme (prenylcysteine methylesterase) prenylcysteine methylesterase (Aes)
PHATRDRAFT_45965 dmt family transporter: drug metabolite (RhaT)
PHATRDRAFT_46363 craniofacial development protein 1 (BCNT)
PHATRDRAFT_46612 mitochondrial carrier protein
PHATRDRAFT_50738 triosephosphate isomerase (TIM)
PHATRDRAFT_6277 lysine decarboxylase (DNA_processg_A superfamily)
PHATRDRAFT_8363 mitochondrial import inner membrane translocase subunit tim10 (zf-Tim10_DDP)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006414 | translational elongation | 0.00164601 | 1 | Phatr_bicluster_0093 |
| GO:0051339 | regulation of lyase activity | 0.00444554 | 1 | Phatr_bicluster_0093 |
| GO:0045039 | mitochondrial inner membrane protein import | 0.0132807 | 1 | Phatr_bicluster_0093 |
| GO:0006626 | protein-mitochondrial targeting | 0.0132807 | 1 | Phatr_bicluster_0093 |
| GO:0016117 | carotenoid biosynthesis | 0.0132807 | 1 | Phatr_bicluster_0093 |
| GO:0009081 | branched chain family amino acid metabolism | 0.0132807 | 1 | Phatr_bicluster_0093 |
| GO:0009073 | aromatic amino acid family biosynthesis | 0.0220418 | 1 | Phatr_bicluster_0093 |
| GO:0000105 | histidine biosynthesis | 0.0393439 | 1 | Phatr_bicluster_0093 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0436241 | 1 | Phatr_bicluster_0093 |
| GO:0008152 | metabolism | 0.137805 | 1 | Phatr_bicluster_0093 |

Comments