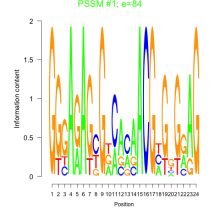
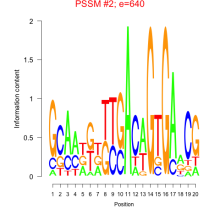
Phatr_bicluster_0149 Residual: 0.22
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0149 | 0.22 | Phaeodactylum tricornutum |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_2171 nitrate transporter (2A0108)
PHATRDRAFT_24019 transcriptional co-repressor sin3a isoform 6 (Sin3_corepress)
PHATRDRAFT_34216 phosducin family protein (Thioredoxin_like superfamily)
PHATRDRAFT_34385 peptidylprolyl isomerase fk506-binding protein (FKBP_C)
PHATRDRAFT_43115 ring-15 protein (RING)
PHATRDRAFT_43375 (zf-C3HC superfamily)
PHATRDRAFT_43880 (LRR_RI superfamily)
PHATRDRAFT_44536 proteophosphoglycan 5
PHATRDRAFT_45955 phospholipase carboxylesterase familyexpressed (COG1054)
PHATRDRAFT_46214 dienelactone hydrolase (COG0412)
PHATRDRAFT_46360 uncharacterised protein family containingexpressed (Bestrophin superfamily)
PHATRDRAFT_46578 tocopherol cyclase
PHATRDRAFT_47127 atp-dependent dna helicase (KU superfamily)
PHATRDRAFT_47879 (SH2_2)
PHATRDRAFT_48345 phosphodiesterasecalmodulin-dependentisoform cra_a
PHATRDRAFT_9278 histone deacetylase superfamily (HDAC_classIV)
PHATRDRAFT_9612 ribose phosphate pyrophosphokinase (PLN02297)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006303 | double-strand break repair via nonhomologous end-joining | 0.00394818 | 1 | Phatr_bicluster_0149 |
| GO:0009165 | nucleotide biosynthesis | 0.00788271 | 1 | Phatr_bicluster_0149 |
| GO:0009116 | nucleoside metabolism | 0.0176596 | 1 | Phatr_bicluster_0149 |
| GO:0006520 | amino acid metabolism | 0.0464863 | 1 | Phatr_bicluster_0149 |
| GO:0016567 | protein ubiquitination | 0.13382 | 1 | Phatr_bicluster_0149 |
| GO:0006457 | protein folding | 0.209255 | 1 | Phatr_bicluster_0149 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.359328 | 1 | Phatr_bicluster_0149 |
| GO:0006508 | proteolysis and peptidolysis | 0.365994 | 1 | Phatr_bicluster_0149 |

Comments