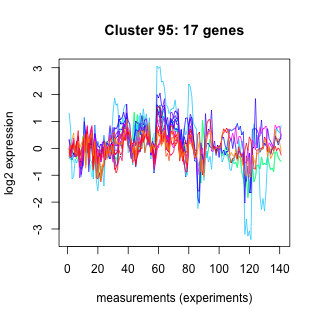
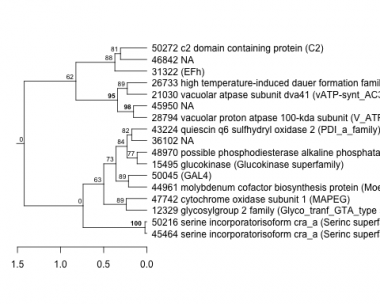
Phatr_hclust_0095 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006777 | Mo-molybdopterin cofactor biosynthesis | 0.00886367 | 1 | Phatr_hclust_0095 |
| GO:0015992 | proton transport | 0.02058 | 1 | Phatr_hclust_0095 |
| GO:0006096 | glycolysis | 0.0507874 | 1 | Phatr_hclust_0095 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0536233 | 1 | Phatr_hclust_0095 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.283834 | 1 | Phatr_hclust_0095 |
| GO:0006118 | electron transport | 0.294987 | 1 | Phatr_hclust_0095 |
|
PHATRDRAFT_45464 : serine incorporatorisoform cra_a (Serinc superfamily) |
PHATRDRAFT_44961 : molybdenum cofactor biosynthesis protein (MoeA) |
PHATRDRAFT_21030 : vacuolar atpase subunit dva41 (vATP-synt_AC39 superfamily) |
|
|
PHATRDRAFT_50216 : serine incorporatorisoform cra_a (Serinc superfamily) |
PHATRDRAFT_50045 : (GAL4) |
PHATRDRAFT_43224 : quiescin q6 sulfhydryl oxidase 2 (PDI_a_family) |
PHATRDRAFT_31322 : (EFh) |
|
PHATRDRAFT_12329 : glycosylgroup 2 family (Glyco_tranf_GTA_type superfamily) |
PHATRDRAFT_15495 : glucokinase (Glucokinase superfamily) |
PHATRDRAFT_28794 : vacuolar proton atpase 100-kda subunit (V_ATPase_I) |
|
|
PHATRDRAFT_47742 : cytochrome oxidase subunit 1 (MAPEG) |
PHATRDRAFT_48970 : possible phosphodiesterase alkaline phosphataseof possible plant or bacterial origin (MPP_PhoD) |
PHATRDRAFT_50272 : c2 domain containing protein (C2) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.810 | 0.000505051 |
| Copper_SH | Copper_SH | 0.164 | 0.150627 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.393 | 0.0201923 |
| Silver_SH | Silver_SH | -0.174 | 0.102888 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.135 | 0.0938849 |
| BlueLight_24h | BlueLight_24h | -0.079 | 0.783837 |
| GreenLight_0.5h | GreenLight_0.5h | 0.733 | 0.0633333 |
| RedLight_6h | RedLight_6h | 0.924 | 0.000694444 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 1 |
| GreenLight_24h | GreenLight_24h | -0.055 | 0.813247 |
| light_6hr | light_6hr | 0.161 | 0.60323 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | -0.127 | 0.627621 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.085 | 0.493004 |
| Ammonia_SH | Ammonia_SH | -0.534 | 0.0863636 |
| Simazine_SH | Simazine_SH | 0.211 | 0.114399 |
| Oil_SH | Oil_SH | -0.091 | 0.414062 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | -0.216 | 0.0987273 |
| Cadmium_SH | Cadmium_SH | 0.251 | 0.0685714 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.417 | 0.0357143 |
| highlight_0to6h | highlight_0to6h | -0.002 | 0.987675 |
| BlueLight_0.5h | BlueLight_0.5h | -0.164 | 0.797307 |
| Dispersant_SH | Dispersant_SH | -0.084 | 0.464835 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | 0.497 | 0.00461539 |
| light_15.5hr | light_15.5hr | 0.604 | 0.0319355 |
| light_10.5hr | light_10.5hr | 0.459 | 0.124558 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.168 | 0.160774 |
| Si_free | Si_free | -0.078 | 0.520879 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 0.391 | 0.470642 |
| Mixture_SH | Mixture_SH | -1.134 | 0.000675676 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | -0.259 | 0.261859 |
| light_16hr_dark_30min | light_16hr_dark_30min | 0.638 | 0.0213178 |
| BlueLight_6h | BlueLight_6h | 0.665 | 0.00148515 |
| highlight_12to48h | highlight_12to48h | -0.267 | 0.225962 |
| RedLight_24h | RedLight_24h | 0.338 | 0.059854 |
| Dark_treated | Dark_treated | 0.066 | 0.940481 |
| GreenLight_6h | GreenLight_6h | 0.750 | 0.00111111 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.055 | 0.257941 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.024 | 0.832449 |
| RedLight_0.5h | RedLight_0.5h | 1.154 | 0.00318182 |
| Green_vs_Red_6h | Green_vs_Red_6h | -0.174 | 0.44838 |
| Re-illuminated_24h | Re-illuminated_24h | 0.274 | 0.0585938 |