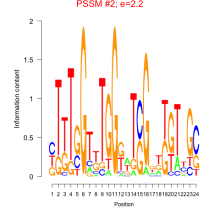
Thaps_bicluster_0194 Residual: 0.41
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0194 | 0.41 | Thalassiosira pseudonana |
Displaying 1 - 36 of 36
" class="views-fluidgrid-wrapper clear-block">
10140 hypothetical protein
10419 hypothetical protein
10485 hypothetical protein
10895 PLN02852
11081 hypothetical protein
11752 2OG-FeII_Oxy_3
1615 hypothetical protein
21141 RibD
22186 MFS
22245 hypothetical protein
24201 hypothetical protein
2426 MviM
24295 (RPN2) RPN2
24529 hypothetical protein
24714 hypothetical protein
24990 hypothetical protein
25158 hypothetical protein
25168 hypothetical protein
25225 Cullin_binding superfamily
25263 Patatin_and_cPLA2 superfamily
25360 hypothetical protein
260758 TFCD_C
262298 BaeS
263118 ABC_ATPase superfamily
263349 COG4178
263989 (HSF13) hypothetical protein
264064 SbcC
31658 DUF3437 superfamily
38294 CBS_like
5079 TRM13 superfamily
5105 hypothetical protein
6170 hypothetical protein
7363 hypothetical protein
7569 hypothetical protein
7886 hypothetical protein
8159 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009231 | riboflavin biosynthesis | 0.0184724 | 1 | Thaps_bicluster_0194 |
| GO:0016310 | phosphorylation | 0.0396209 | 1 | Thaps_bicluster_0194 |
| GO:0015992 | proton transport | 0.0455847 | 1 | Thaps_bicluster_0194 |
| GO:0006520 | amino acid metabolism | 0.0632695 | 1 | Thaps_bicluster_0194 |
| GO:0007018 | microtubule-based movement | 0.125531 | 1 | Thaps_bicluster_0194 |
| GO:0008152 | metabolism | 0.126308 | 1 | Thaps_bicluster_0194 |
| GO:0000074 | regulation of cell cycle | 0.170973 | 1 | Thaps_bicluster_0194 |
| GO:0006118 | electron transport | 0.198115 | 1 | Thaps_bicluster_0194 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.245811 | 1 | Thaps_bicluster_0194 |
| GO:0006468 | protein amino acid phosphorylation | 0.491617 | 1 | Thaps_bicluster_0194 |

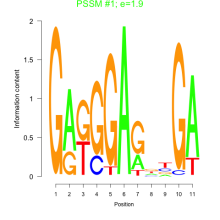
Comments