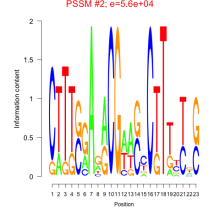
Phatr_bicluster_0218 Residual: 0.32
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0218 | 0.32 | Phaeodactylum tricornutum |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_13894 dihydrolipoamide s-acetyltransferase precursor (PDHac_trf_mito)
PHATRDRAFT_29016 2-oxoglutarate dehydrogenase e1 oxoglutarate alpha-ketoglutaric (sucA)
PHATRDRAFT_33218 (ArsB_NhaD_permease superfamily)
PHATRDRAFT_36398 agmatine deiminase (PLN02690)
PHATRDRAFT_38362 2-isopropylmalate synthase (PRK12344)
PHATRDRAFT_39438 loc779553 protein
PHATRDRAFT_43090 methyltransferase type 11
PHATRDRAFT_47510 aldo keto reductase-like protein (Aldo_ket_red)
PHATRDRAFT_47517 gyltl1b protein (Glyco_transf_49 superfamily)
PHATRDRAFT_48603 amineflavin-containing superfamily (Amino_oxidase)
PHATRDRAFT_48955 (Cation_efflux superfamily)
PHATRDRAFT_49115 (AlgLyase superfamily)
PHATRDRAFT_49734 protein familywith a coiled coil-4 domain (Methyltransf_22)
PHATRDRAFT_605 mitochondrial helicase twinkle (RecA-like_NTPases superfamily)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019752 | carboxylic acid metabolism | 0.00444115 | 1 | Phatr_bicluster_0218 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0350414 | 1 | Phatr_bicluster_0218 |
| GO:0006812 | cation transport | 0.058495 | 1 | Phatr_bicluster_0218 |
| GO:0015031 | protein transport | 0.066897 | 1 | Phatr_bicluster_0218 |
| GO:0008152 | metabolism | 0.134877 | 1 | Phatr_bicluster_0218 |
| GO:0006412 | protein biosynthesis | 0.306181 | 1 | Phatr_bicluster_0218 |
| GO:0006508 | proteolysis and peptidolysis | 0.401138 | 1 | Phatr_bicluster_0218 |
| GO:0006118 | electron transport | 0.408154 | 1 | Phatr_bicluster_0218 |


Comments