Using the TFOE transcriptional regulation network spreadsheet tool
Tige Rustad, Senior Scientist at Seattle Biomed
Email: tige.rustad@seattlebiomed.org
What is the TFOE network?
- TFOE- transcription factor over expression
- 206 transcription factors (TFs) cloned into an inducible tagged expression vector
- Microarrays to identify genes differentialy expressed (DE)
DE =the set of genes repressed OR induced
- ChIP-seq to identify DNA bound by each TF
- These two parallel approaches were published in:
Rustad & Minch et al. 2014. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biology 15:502
Minch & Rustad et al. 2014. The DNA binding network of Mycobacterium tuberculosis. Nature Communications
What does the spreadsheet do?
- Users input a gene or list of genes of interest (GOI). The spreadsheet then:
- Identifies all TFs that alter expression of any of your GOI
- Shows how many of your GOI are altered by each TF
- Calculates the significance of the overlap between your list of genes and the set of genes altered by each TF
- Identifies TFs that bind in upstream of the GOI
Getting started
- Download and open the most recent version of the spreadsheet at :
http://networks.systemsbiology.net/mtb/content/TFOE-Searchable-Data-File
- The spreadsheet should open to the tab 'ListQuery'
- The spreadsheet consists of six tabs:
ListQuery - input GOI and see an overview of results TFOE.Readout - Expression changes in your GOI for each TFChIP.Readout - DNA binding proximal to your GOITFOE.data - Expression changes triggered by TFOE and p-value of each changeChIP.data - For each gene indicates which TFs bind directly upstream or upstream of gene earlier in the operonAnnot - Gene annotation pulled from Tuberculist and PATRIC
Step One: Choose your gene(s) of interest

Choose your genes of interest- at least one, up to 1,000
- For this example we will use the 101 reaeration induced genes from Sherrid et al.
You can access this list here: Reaeration Response.xlsx
- Genes need to be in the format from the original sequencing of the H37Rv geneome (i.e. "Rv####" with a terminal 'c' if the gene is on the Crick strand)
Step Two: Input your genes of interest (GOI)

Paste your genes into the blue cells
#TFOE is the number of TFs that change expression that gene- You can change the
fold change and p-value considered significant by changing the values in the red cells. Default is log2 of 1 (two fold) and pvalue ≤ 0.01
- The total number of genes in your list and the number of those genes that are perturbed by at least one TF are indicated
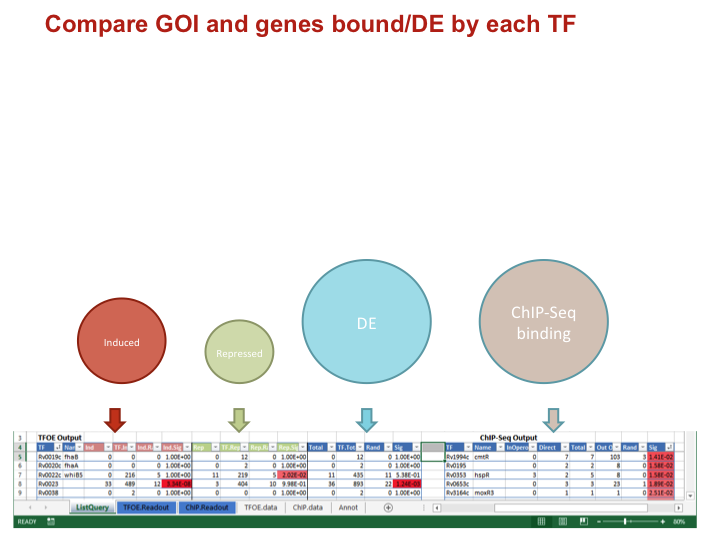
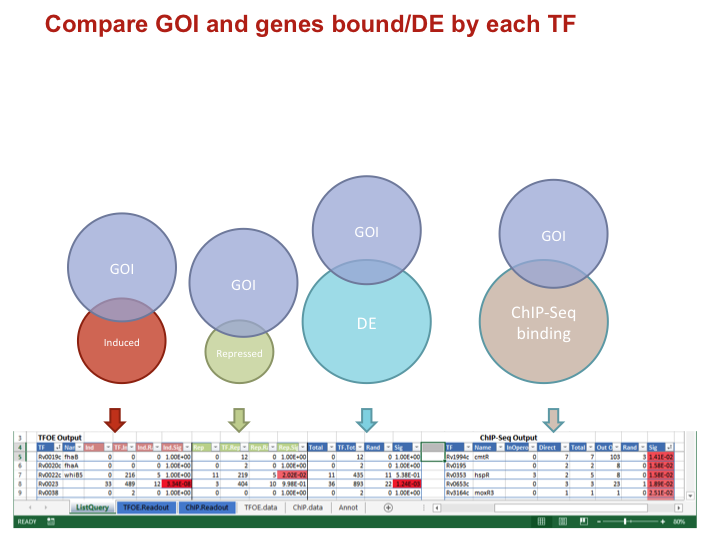
Identify overlaps between GOI and each TF
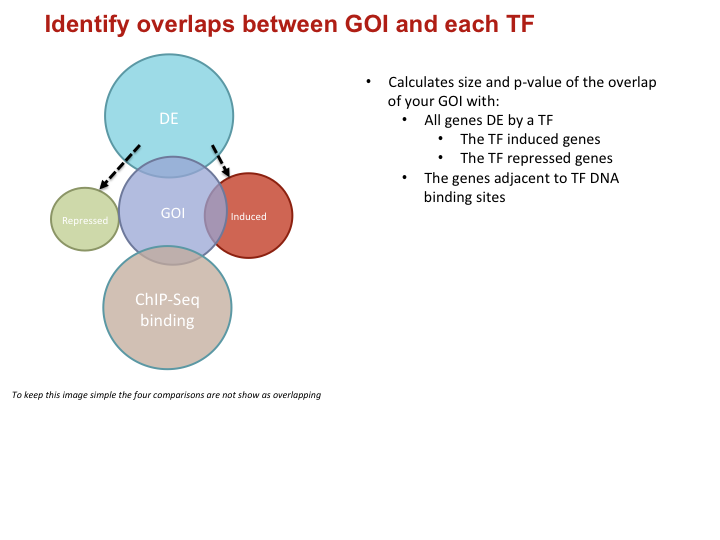
- Calculates size and p-value of the overlap of your GOI with:
- All genes DE by a TF
- The TF induced genes
- The TF repressed genes
- The genes adjacent to TF DNA binding sites
To keep this image simple the four comparisons are not show as overlapping
Compare GOI and genes bound/DE by each TF
Data types for each comparison
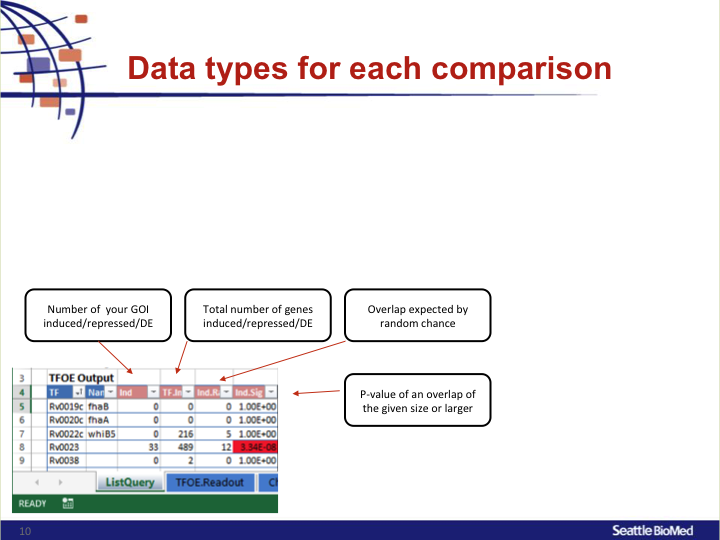
IndNumber of your GOI induced/repressed/DETF.IndTotal number of genes induced/repressed/DEInd.RandOverlap expected by random chanceInd.SigP-value of an overlap of the given size or larger
Step Three: Sort the output

Sort the TFs as desired
Each column title has an arrow that pulls up sorting options
- Sorting by the significance or size of the overlap is generally the most useful view
- The order of TFs in the Output table determines the order of TFs in the 'Readout' tabs
Step Four: TFOE.Readout
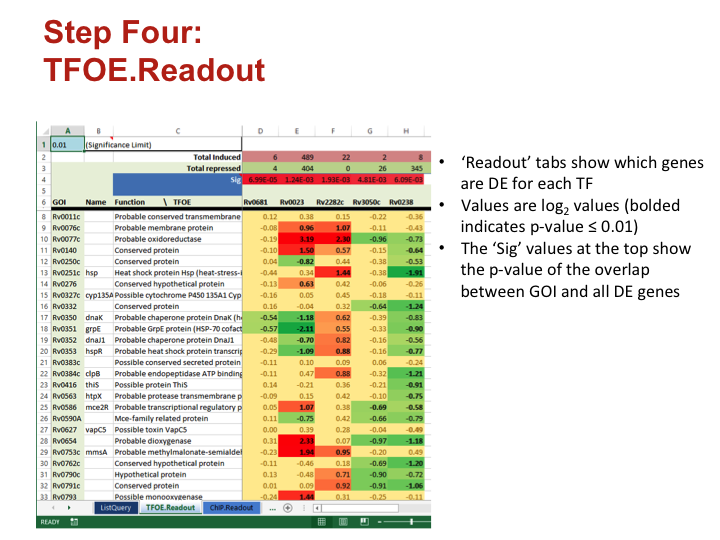
- ‘Readout’ tabs show which genes are DE for each TF
- Values are log2 values (bolded indicates p-value ≤ 0.01)
- The ‘Sig’ values at the top show the p-value of the overlap between GOI and all DE genes