Data Center
MTB Signed TRN
MTB Signed Transcriptional Regulatory Network
The following file is a supplementary material for Peterson et al.(submitted)
The compiled signed transcriptional regulatory network contains TF-gene interactions with both physical (detected with ChIP-seq experiments; Minch et al. 2015) and functional evidence (detected with TFOE transcriptional profiling; Rustad et al. 2014). Activation and repression are indicated with 1 and -1.
OTB supplement: Systems analysis of TB progression in RhCMV/MTB vaccinated NHP. Whole blood multi-omics profiling of MTB-challenged Rhesus macaques after vaccination with RhCMV and/or BCG.
Currently, data files in this section are private and download link will be available after login.
| Title | Body | Release Date | Sample Type |
|---|---|---|---|
| RhCMV-TB Metabolomics Data |
Whole blood metabolomics profiling of MTB-challenged Rhesus macaques after vaccination with RhCMV and/or BCG. Each row is a metabolite and each column is a monkey at the defined time point |
2018 | Metabolomics |
| RhCMV-TB Proteomics Data |
Whole blood proteomics profiling of MTB-challenged Rhesus macaques after vaccination with RhCMV and/or BCG |
2018 | Protein |
| RhCMV-TB Transcriptomic Data |
Whole blood transcriptomes of MTB-challenged Rhesus macaques after vaccination with RhCMV and/or BCG Log2 standardized counts (using edgeR) are reported. Each row is a gene and each column is a monkey at the defined time point. |
2018 | RNA |
| RhCMV-TB Meta Data |
Whole blood omics profiling of MTB-challenged Rhesus macaques after vaccination with RhCMV and/or BCG Meta-data information is listed. Columns are the monkey ID, which trial the monkey is from, the disease score after challenge, and the vaccination group. |
2018 | Meta-data |
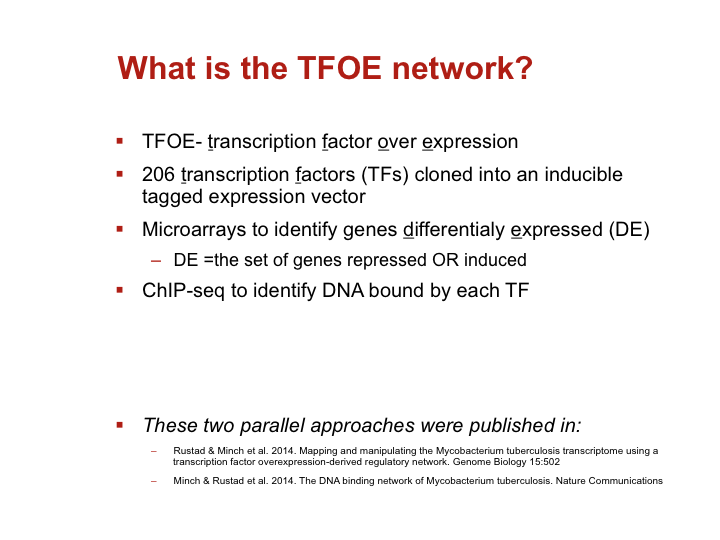
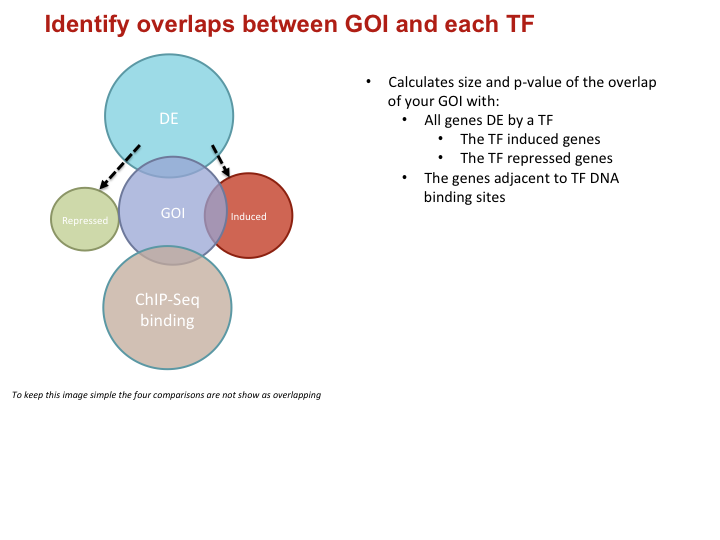
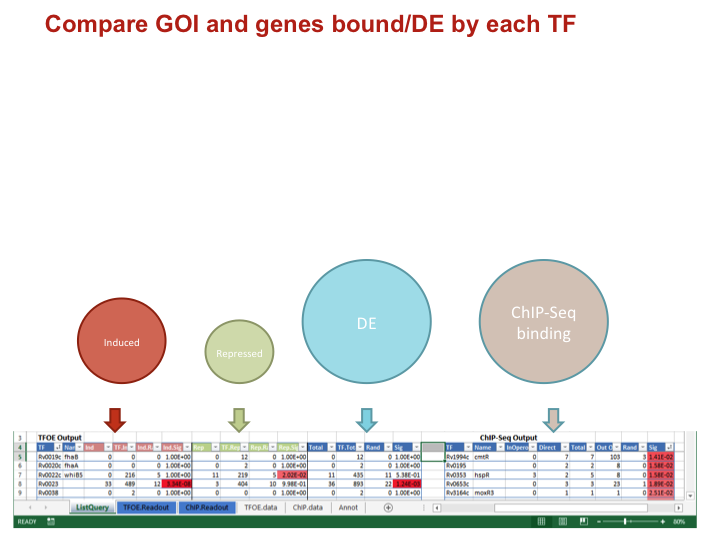
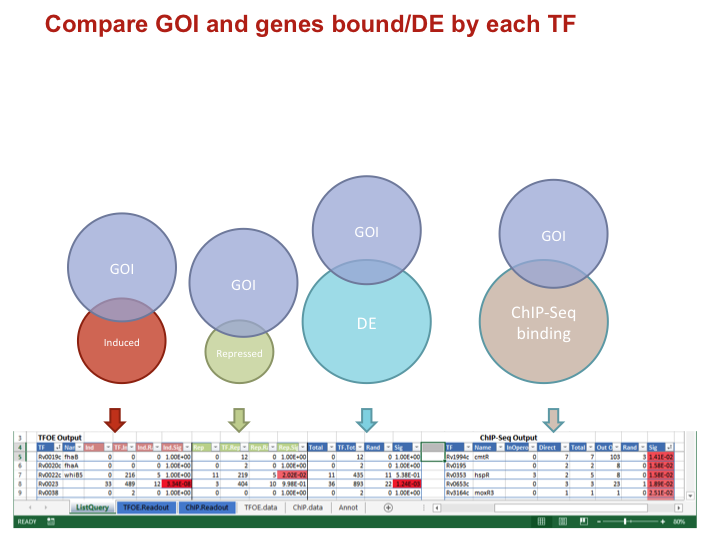
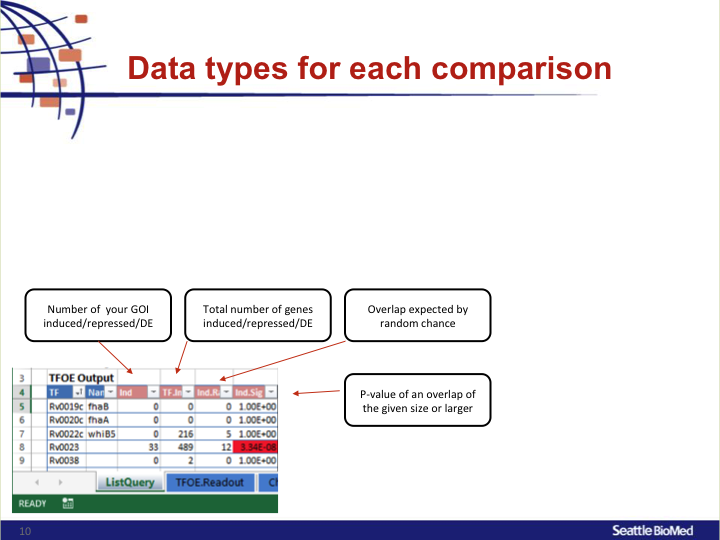
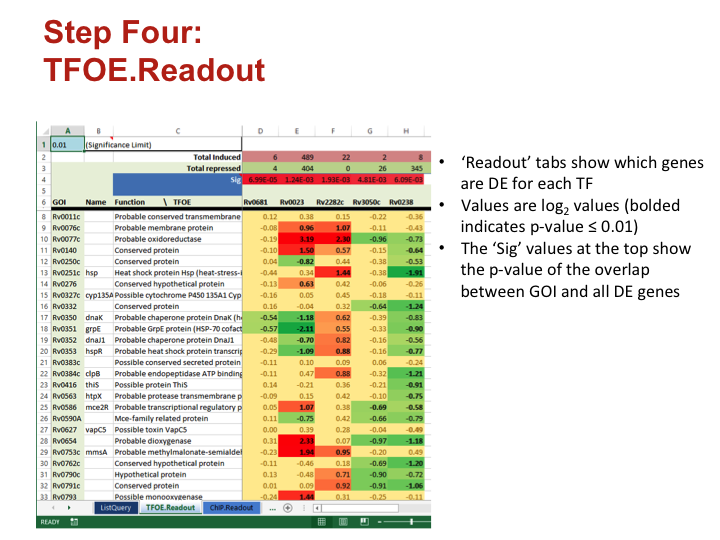
To investigate the MTB transcriptional landscape in a systematic manner, we developed a high-throughput approach to identify the genes controlled by nearly all predicted MTB TFs. We individually cloned and conditionally overexpressed 206 MTB TFs to induce the regulatory signature of each one. Using this approach we identified the sets of genes affected by TF overexpression (TFOE) and assembled them into an easily searchable map of transcriptional regulation in MTB.
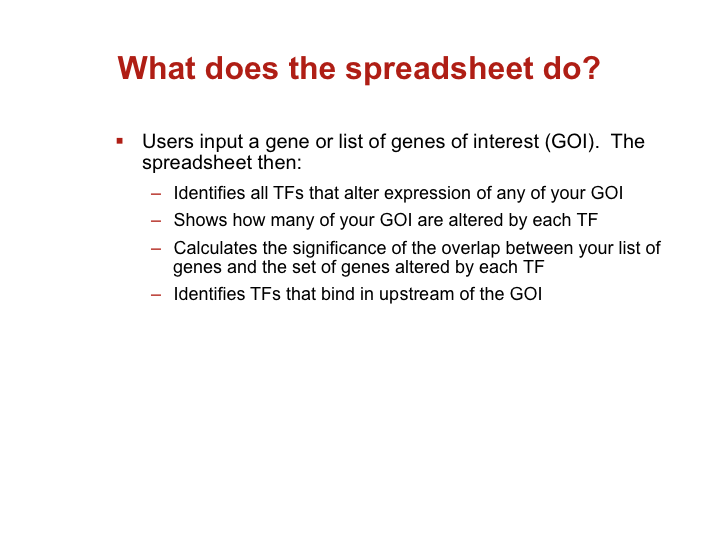
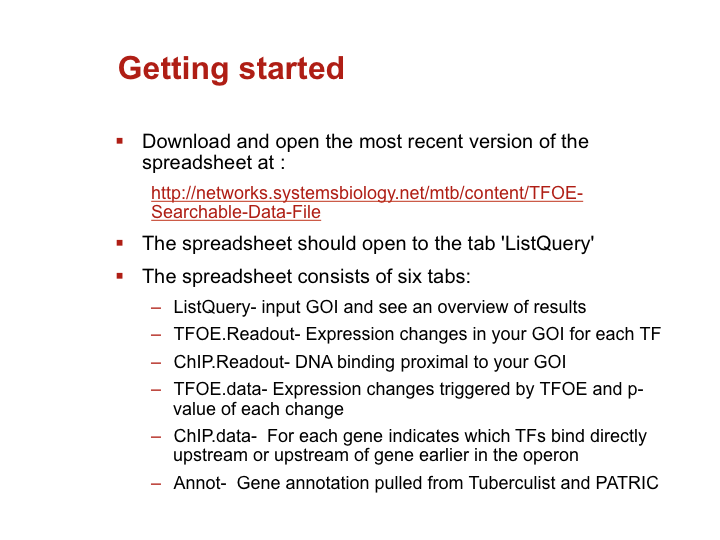
Accessing large datasets like the TFOE expression data can be difficult when the data spreads over thousands of genes and hundreds of regulators. To address the difficulties usually associated with accessing large data sets, we have designed a simple Excel spreadsheet for querying TFOE data to find regulators of specific genes or sets of genes. (Rustad et al. Genome Biol. 2014)
Last update: 29/02/2015
Whole blood metabolomics profiling of MTB-challenged Rhesus macaques after vaccination with RhCMV and/or BCG. Each row is a metabolite and each column is a monkey at the defined time point
| Title | Gene | BioProject | GEO Series | Platform | Accession | Sample Method | Sample Type | References | Release Date | Repository | Contact |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RhCMV-TB Metabolomics Data | Metabolomics | 2018 | |||||||||
| RhCMV-TB Proteomics Data | Protein | 2018 | |||||||||
| RhCMV-TB Transcriptomic Data | RNA-seq | RNA | 2018 | ||||||||
| RhCMV-TB Meta Data | in progress | NA | NA | Meta-data | in preparation | 2018 | in progress | ||||
| TFOE_8468_3911 | PRJNA254351 | GSE59086 | GPL14824 | GSM1427062 | Tiling Array | RNA | 25232098 | 4-Jul-14 | GEO | ||
| TFOE_8127_3911 | PRJNA254351 | GSE59086 | GPL14824 | GSM1427061 | Tiling Array | RNA | 25232098 | 4-Jul-14 | GEO | ||
| TFOE_2756_3911 | PRJNA254351 | GSE59086 | GPL14824 | GSM1427060 | Tiling Array | RNA | 25232098 | 4-Jul-14 | GEO | ||
| TFOE_7945_3862c_B | PRJNA254351 | GSE59086 | GPL14824 | GSM1427059 | Tiling Array | RNA | 25232098 | 4-Jul-14 | GEO | ||
| TFOE_6830_3862c | PRJNA254351 | GSE59086 | GPL14824 | GSM1427058 | Tiling Array | RNA | 25232098 | 4-Jul-14 | GEO | ||
| TFOE_4985_3862c_A | PRJNA254351 | GSE59086 | GPL14824 | GSM1427057 | Tiling Array | RNA | 25232098 | 4-Jul-14 | GEO |