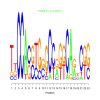
Module 145 Residual: 0.56
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0145 | v02 | 0.56 | -16.64 |
| Rv0944 Formamidopyrimidine-DNA glycosylase (EC 3.2.2.23) | Rv0945 Oxidoreductase, short-chain dehydrogenase/reductase family (EC 1.1.1.-) |
| Rv0955 FIG021574: Possible membrane protein related to de Novo purine biosynthesis | Rv1010 Dimethyladenosine transferase (EC 2.1.1.-) |
| Rv1415 3,4-dihydroxy-2-butanone 4-phosphate synthase / GTP cyclohydrolase II (EC 3.5.4.25) | Rv1416 6,7-dimethyl-8-ribityllumazine synthase (EC 2.5.1.78) |
| Rv1417 Cell division protein DivIC (FtsB), stabilizes FtsL against RasP cleavage | Rv1897c D-tyrosyl-tRNA(Tyr) deacylase |
| Rv1898 ExtraCellular Mutant; Ecm15p | Rv2772c |
| Rv2849c Cob(I)alamin adenosyltransferase (EC 2.5.1.17) | Rv2850c ChlI component of cobalt chelatase involved in B12 biosynthesis / ChlD component of cobalt chelatase involved in B12 biosynthesis |
| Rv3592 Antibiotic biosynthesis monooxygenase | Rv3593 Beta-lactamase (EC 3.5.2.6) |
| Rv0953c Hydride transferase 1 (Fragment) | Rv0954 antigen 34 kDa |
| Module GO Term Enrichment | Descriptions |
|---|---|
| GO:0006771 | GO:0009231 | GO:0042726 | GO:0042727 | riboflavin metabolic process | riboflavin biosynthetic process | flavin-containing compound metabolic pro... | flavin-containing compound biosynthetic ... |
|
GO:0004519 | endonuclease activity |
|
GO:0005886 | plasma membrane |
|
GO:0006281 | DNA repair |
|
GO:0006289 | nucleotide-excision repair |
|
GO:0034599 | cellular response to oxidative stress |
|
GO:0040007 | growth |
|
GO:0008649 | rRNA methyltransferase activity |
|
GO:0000154 | rRNA modification |
|
GO:0005576 | extracellular region |
|
GO:0004746 | riboflavin synthase activity |
|
GO:0019478 | D-amino acid catabolic process |
|
GO:0005737 | cytoplasm |
|
GO:0008817 | cob(I)yrinic acid a,c-diamide adenosyltransferase activity |
|
GO:0005887 | integral to plasma membrane |


















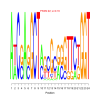
Discussion