Module 587 Residual: 0.58
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0587 | v02 | 0.58 | -11.83 |
| Rv1112 GTP-binding and nucleic acid-binding protein YchF | Rv0787 |
| Rv2191 DNA polymerase III epsilon subunit (EC 2.7.7.7) | Rv2296 Haloalkane dehalogenase (EC 3.8.1.5) |
| Rv2423 SAM-dependent methyltransferases | Rv2447c Dihydrofolate synthase (EC 6.3.2.12) / Folylpolyglutamate synthase (EC 6.3.2.17) |
| Rv2469c HNH endonuclease family protein | Rv2470 Hemoglobin-like protein HbO |
| Rv2471 Alpha-glucosidase (EC 3.2.1.20) | Rv2853 PE-PGRS FAMILY PROTEIN |
| Rv3109 | Rv3110 Pterin-4-alpha-carbinolamine dehydratase (EC 4.2.1.96) |
| Rv3111 Molybdenum cofactor biosynthesis protein MoaC | Rv3112 Molybdenum cofactor biosynthesis protein MoaD |
| Rv3309c Xanthine-guanine phosphoribosyltransferase (EC 2.4.2.22) | Rv3310 Possible acid phosphatase (EC 3.1.3.2) |
| Rv3905c Esat-6 like protein EsxF |
|
GO:0005622 | intracellular |
|
GO:0005525 | GTP binding |
|
GO:0005886 | plasma membrane |
|
GO:0005576 | extracellular region |
|
GO:0003887 | DNA-directed DNA polymerase activity |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0000287 | magnesium ion binding |
|
GO:0040007 | growth |
|
GO:0043531 | ADP binding |
|
GO:0050897 | cobalt ion binding |
|
GO:0015671 | oxygen transport |
|
GO:0019825 | oxygen binding |
|
GO:0020037 | heme binding |
|
GO:0042803 | protein homodimerization activity |
|
GO:0051260 | protein homooligomerization |
|
GO:0006777 | Mo-molybdopterin cofactor biosynthetic process |
|
GO:0006790 | sulfur compound metabolic process |
|
GO:0004845 | uracil phosphoribosyltransferase activity |
|
GO:0003993 | acid phosphatase activity |
|
GO:0004438 | phosphatidylinositol-3-phosphatase activity |
|
GO:0004805 | trehalose-phosphatase activity |
|
GO:0006184 | GTP catabolic process |
|
GO:0006742 | NADP catabolic process |
|
GO:0046854 | phosphatidylinositol phosphorylation |
|
GO:0050189 | phosphoenolpyruvate phosphatase activity |
|
GO:0050192 | phosphoglycerate phosphatase activity |



















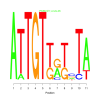
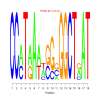
Discussion