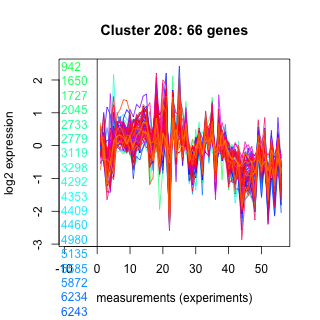
Thaps_hclust_0208 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006464 | protein modification | 0.00502878 | 1 | Thaps_hclust_0208 |
| GO:0019363 | pyridine nucleotide biosynthesis | 0.0119562 | 1 | Thaps_hclust_0208 |
| GO:0046168 | glycerol-3-phosphate catabolism | 0.0178824 | 1 | Thaps_hclust_0208 |
| GO:0009435 | NAD biosynthesis | 0.0178824 | 1 | Thaps_hclust_0208 |
| GO:0006512 | ubiquitin cycle | 0.0192775 | 1 | Thaps_hclust_0208 |
| GO:0006801 | superoxide metabolism | 0.0237743 | 1 | Thaps_hclust_0208 |
| GO:0006072 | glycerol-3-phosphate metabolism | 0.0237743 | 1 | Thaps_hclust_0208 |
| GO:0001539 | ciliary or flagellar motility | 0.0237743 | 1 | Thaps_hclust_0208 |
| GO:0006207 | 'de novo' pyrimidine base biosynthesis | 0.0296321 | 1 | Thaps_hclust_0208 |
| GO:0018346 | protein amino acid prenylation | 0.0296321 | 1 | Thaps_hclust_0208 |
|
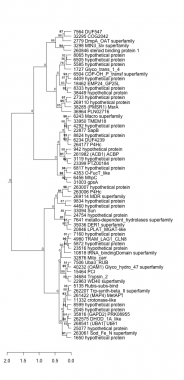
942 : hypothetical protein |
6243 : Macro superfamily |
18462 : EMP24_GP25L |
36964 : PLN02716 |
|
1650 : hypothetical protein |
6456 : MhpC |
20846 : LPLAT_MGAT-like |
38285 : (PMSR1) MsrA |
|
1727 : Glyco_trans_1_4 |
6504 : CDP-OH_P_transf superfamily |
22877 : SapB |
38449 : hypothetical protein |
|
2045 : hypothetical protein |
6505 : hypothetical protein |
22963 : WD40 superfamily |
40232 : (OAM1) Glyco_hydro_47 superfamily |
|
2733 : hypothetical protein |
6817 : hypothetical protein |
23399 : PTZ00184 |
260846 : steroid binding protein 1 |
|
2779 : DmpA_OAT superfamily |
7160 : hypothetical protein |
23516 : hypothetical protein |
261422 : (MAP4) MetAP1 |
|
3119 : hypothetical protein |
7506 : Uba3_RUB |
24754 : hypothetical protein |
261982 : (ACB1) ACBP |
|
3298 : MtN3_slv superfamily |
7564 : DUF547 |
25077 : hypothetical protein |
262207 : Trp-synth-beta_II superfamily |
|
4292 : hypothetical protein |
7641 : metallo-dependent_hydrolases superfamily |
31003 : gpsA |
262575 : DHOD_1A_like |
|
4353 : O-FucT_like |
8065 : hypothetical protein |
32295 : COG2042 |
263006 : P4Hc |
|
4409 : hypothetical protein |
8333 : hypothetical protein |
32876 : Mito_carr |
263007 : hypothetical protein |
|
4460 : hypothetical protein |
8599 : hypothetical protein |
33094 : Sun |
263061 : Sod_Fe_N superfamily |
|
4980 : TRAM_LAG1_CLN8 |
8824 : hypothetical protein |
33950 : TMEM18 |
264177 : P4Hc |
|
5135 : Rubis-subs-bind |
9834 : hypothetical protein |
34684 : Trypsin_2 |
268541 : (UBA1) Ube1 |
|
5585 : hypothetical protein |
10818 : tRNA_bindingDomain superfamily |
35036 : DER1 superfamily |
269110 : hypothetical protein |
|
5872 : hypothetical protein |
11332 : crotonase-like |
35816 : (GAPD2) PRK08955 |
269114 : MDR superfamily |
|
6234 : DUF4239 |
15464 : PCI |
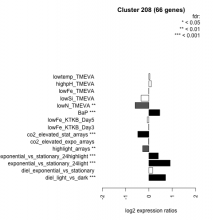
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.911 | 0.000581 |
| diel_light_vs_dark | diel_light_vs_dark | 0.714 | 0.000485 |
| BaP | BaP | 0.466 | 0.00037 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.404 | 0.000526 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.161 | 0.099 |
| highpH_TMEVA | highpH_TMEVA | 0.102 | 0.329 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.044 | 0.8 |
| lowFe_TMEVA | lowFe_TMEVA | -0.004 | 0.996 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.007 | 0.968 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.014 | 0.985 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.072 | 0.561 |
| highlight_arrays | highlight_arrays | -0.269 | 0.00229 |
| lowSi_TMEVA | lowSi_TMEVA | -0.334 | 0.445 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.474 | 0.000658 |
| lowN_TMEVA | lowN_TMEVA | -0.590 | 0.00119 |