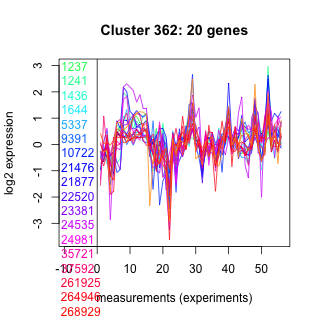
Thaps_hclust_0362 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019363 | pyridine nucleotide biosynthesis | 0.00371824 | 1 | Thaps_hclust_0362 |
| GO:0006825 | copper ion transport | 0.00557274 | 1 | Thaps_hclust_0362 |
| GO:0017000 | antibiotic biosynthesis | 0.00927255 | 1 | Thaps_hclust_0362 |
| GO:0006364 | rRNA processing | 0.0147993 | 1 | Thaps_hclust_0362 |
| GO:0006629 | lipid metabolism | 0.0527255 | 1 | Thaps_hclust_0362 |
| GO:0006468 | protein amino acid phosphorylation | 0.0566487 | 1 | Thaps_hclust_0362 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.101273 | 1 | Thaps_hclust_0362 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.446038 | 1 | Thaps_hclust_0362 |
|
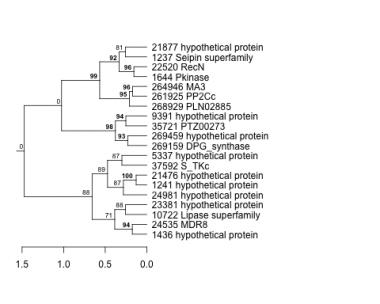
1237 : Seipin superfamily |
9391 : hypothetical protein |
23381 : hypothetical protein |
261925 : PP2Cc |
|
1241 : hypothetical protein |
10722 : Lipase superfamily |
24535 : MDR8 |
264946 : MA3 |
|
1436 : hypothetical protein |
21476 : hypothetical protein |
24981 : hypothetical protein |
268929 : PLN02885 |
|
1644 : Pkinase |
21877 : hypothetical protein |
35721 : PTZ00273 |
269159 : DPG_synthase |
|
5337 : hypothetical protein |
22520 : RecN |
37592 : S_TKc |
269459 : hypothetical protein |
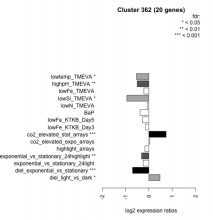
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.477 | 0.0166 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.136 | 0.604 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.275 | 0.171 |
| BaP | BaP | -0.379 | 0.0779 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.314 | 0.0013 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.737 | 0.000658 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.535 | 0.0382 |
| highpH_TMEVA | highpH_TMEVA | -0.502 | 0.00309 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.041 | 0.934 |
| lowFe_TMEVA | lowFe_TMEVA | -0.218 | 0.432 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -0.249 | 0.475 |
| lowN_TMEVA | lowN_TMEVA | -0.071 | 0.86 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.700 | 0.000602 |
| lowSi_TMEVA | lowSi_TMEVA | -0.944 | 0.0123 |
| highlight_arrays | highlight_arrays | -0.169 | 0.308 |