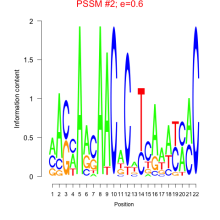
Thaps_bicluster_0005 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0005 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 53 of 53
" class="views-fluidgrid-wrapper clear-block">
1005 hypothetical protein
10275 hypothetical protein
1060 GFA superfamily
1080 hypothetical protein
10805 hypothetical protein
11019 hypothetical protein
11109 Na_H_Exchanger
11415 hypothetical protein
1149 hypothetical protein
1203 PPI_Ypi1
12050 Mem_trans superfamily
14772 AAT_I superfamily
16842 (SAT1) CysE
18071 Peptidase_C48 superfamily
20600 hypothetical protein
20955 hypothetical protein
21342 hypothetical protein
21666 hypothetical protein
22052 hypothetical protein
2250 hypothetical protein
23776 S_TKc
24634 hypothetical protein
25170 hypothetical protein
25335 hypothetical protein
2576 hypothetical protein
25899 hypothetical protein
25915 hypothetical protein
261298 pas-domain protein
262934 hypothetical protein
264928 FG-GAP_2
2663 PLDc_Tdp1_2
269120 (hsp70) PTZ00009
269391 MATE_like superfamily
269400 p450 superfamily
269421 P4Hc
27834 PcnB
34785 prfB
38132 Rhomboid
4189 hypothetical protein
4599 hypothetical protein
5089 Cluap1
5513 hypothetical protein
5859 COG1233
6307 hypothetical protein
6519 LRR_RI superfamily
6779 hypothetical protein
6843 hypothetical protein
7519 hypothetical protein
7767 hypothetical protein
8280 hypothetical protein
9086 (Tp_HSF_1.3i) HSF_DNA-bind superfamily
9892 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006535 | cysteine biosynthesis from serine | 0.0139887 | 1 | Thaps_bicluster_0005 |
| GO:0006415 | translational termination | 0.0209137 | 1 | Thaps_bicluster_0005 |
| GO:0007160 | cell-matrix adhesion | 0.0615124 | 1 | Thaps_bicluster_0005 |
| GO:0006520 | amino acid metabolism | 0.0714116 | 1 | Thaps_bicluster_0005 |
| GO:0007155 | cell adhesion | 0.100523 | 1 | Thaps_bicluster_0005 |
| GO:0019538 | protein metabolism | 0.150166 | 1 | Thaps_bicluster_0005 |
| GO:0006281 | DNA repair | 0.150166 | 1 | Thaps_bicluster_0005 |
| GO:0006396 | RNA processing | 0.185694 | 1 | Thaps_bicluster_0005 |
| GO:0005975 | carbohydrate metabolism | 0.233639 | 1 | Thaps_bicluster_0005 |
| GO:0006118 | electron transport | 0.23943 | 1 | Thaps_bicluster_0005 |

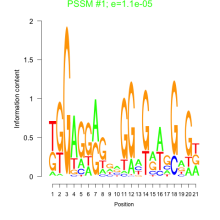
Comments