Thaps_bicluster_0090 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0090 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 31 of 31
" class="views-fluidgrid-wrapper clear-block">
10336 DUF3506 superfamily
1094 hypothetical protein
11564 hypothetical protein
1170 hypothetical protein
1734 hypothetical protein
20564 hypothetical protein
20843 hypothetical protein
20899 (Tp_bHLH2_PAS) regulator [Rayko]
20996 (Tp_bZIP18) regulator [Rayko]
24505 hypothetical protein
24613 Sad1_UNC
24901 (MTS2_2) PRK11893
2537 Patatin_and_cPLA2 superfamily
260900 PRK05347
261633 (CTS2) cysS
263181 cpn10
268070 Trp-synth-beta_II superfamily
2714 hypothetical protein
28865 AarF
3073 SRPBCC superfamily
31265 (CAT1_1) Mito_carr
3215 hypothetical protein
35048 FBP_aldolase_IIA
36929 PCBP_like_KH
3698 (Tp_HSF_1a) HSF_DNA-bind
39606 PRK05431
42971 valS
4632 hypothetical protein
8140 hypothetical protein
8164 hypothetical protein
980 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006418 | tRNA aminoacylation for protein translation | 0.000129917 | 0.356101 | Thaps_bicluster_0090 |
| GO:0030036 | actin cytoskeleton organization and biogenesis | 0.00475501 | 1 | Thaps_bicluster_0090 |
| GO:0016043 | cellular component organization and biogenesis | 0.00475501 | 1 | Thaps_bicluster_0090 |
| GO:0015995 | chlorophyll biosynthetic process | 0.00475501 | 1 | Thaps_bicluster_0090 |
| GO:0006431 | methionyl-tRNA aminoacylation | 0.0094884 | 1 | Thaps_bicluster_0090 |
| GO:0006438 | valyl-tRNA aminoacylation | 0.0094884 | 1 | Thaps_bicluster_0090 |
| GO:0006423 | cysteinyl-tRNA aminoacylation | 0.0142002 | 1 | Thaps_bicluster_0090 |
| GO:0006434 | seryl-tRNA aminoacylation | 0.0142002 | 1 | Thaps_bicluster_0090 |
| GO:0006424 | glutamyl-tRNA aminoacylation | 0.0188906 | 1 | Thaps_bicluster_0090 |
| GO:0006614 | SRP-dependent cotranslational protein-membrane targeting | 0.0282075 | 1 | Thaps_bicluster_0090 |

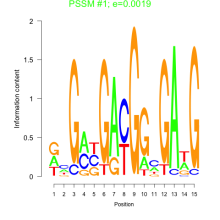
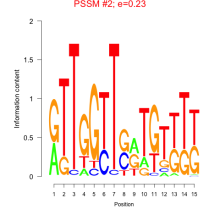
Comments