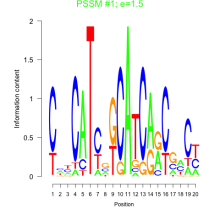
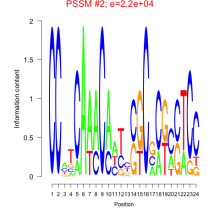
Thaps_bicluster_0126 Residual: 0.34
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0126 | 0.34 | Thalassiosira pseudonana |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
1218 Aa_trans superfamily
1275 hypothetical protein
1741 hypothetical protein
20633 Aa_trans superfamily
20854 hypothetical protein
21238 (ALD3) ALDH_F3-13-14_CALDH-like
21712 APG9 superfamily
22823 Maf1 superfamily
22902 hypothetical protein
23990 hypothetical protein
268867 Peptidase_S28 superfamily
269328 (PCB1) COG4799
269513 PTZ00461
2906 hypothetical protein
31777 (OCT1) CoA_trans superfamily
3260 hypothetical protein
3456 Peptidase_S41_CPP
3493 hypothetical protein
4100 hypothetical protein
4322 PLN00113
5340 hypothetical protein
6047 TLD superfamily
6568 PLPDE_IV superfamily
885 DUF544 superfamily
9861 DUF563
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006865 | amino acid transport | 0.0012175 | 1 | Thaps_bicluster_0126 |
| GO:0006508 | proteolysis and peptidolysis | 0.0134124 | 1 | Thaps_bicluster_0126 |
| GO:0008152 | metabolism | 0.0338556 | 1 | Thaps_bicluster_0126 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0126 |

Comments