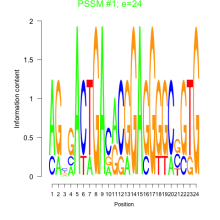
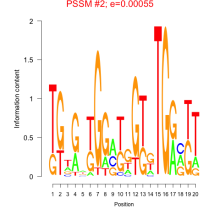
Thaps_bicluster_0174 Residual: 0.29
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0174 | 0.29 | Thalassiosira pseudonana |
Displaying 1 - 19 of 19
" class="views-fluidgrid-wrapper clear-block">
11460 hypothetical protein
1875 hypothetical protein
2039 hypothetical protein
23593 hypothetical protein
261242 (PSBP) PsbP superfamily
261284 Aldo_ket_red
261935 thioredoxin
263116 rubredoxin
263902 PT_UbiA_HPT1
268669 hypothetical protein
2770 hypothetical protein
31294 DUF1230 superfamily
37599 hypothetical protein
3845 hypothetical protein
38769 PLN00033
40966 (Tp_sigma70.6) Sig70-cyanoRpoD
5928 hypothetical protein
7583 DUF1995 superfamily
8293 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006352 | transcription initiation | 0.0133715 | 1 | Thaps_bicluster_0174 |
| GO:0006118 | electron transport | 0.0269761 | 1 | Thaps_bicluster_0174 |
| GO:0015031 | protein transport | 0.0436835 | 1 | Thaps_bicluster_0174 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.27964 | 1 | Thaps_bicluster_0174 |

Comments