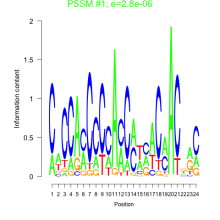
Thaps_bicluster_0175 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0175 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 35 of 35
" class="views-fluidgrid-wrapper clear-block">
20944 Glyco_tranf_GTA_type superfamily
21074 hypothetical protein
21109 DAGK_cat
21255 SYLF
21444 hypothetical protein
24644 hypothetical protein
2516 hypothetical protein
261589 S49_Sppa_N_C
270302 hypothetical protein
2792 hypothetical protein
2816 hypothetical protein
34042 Abhydrolase_6
34077 ISN1 superfamily
36502 IF4E
37791 LanC_like superfamily
38632 Esterase_lipase superfamily
3933 hypothetical protein
3966 PT_UbiA superfamily
4512 hypothetical protein
4650 hypothetical protein
5182 hypothetical protein
5977 hypothetical protein
6089 hypothetical protein
6373 hypothetical protein
6452 hypothetical protein
6468 hypothetical protein
6585 MATH
7085 PI-PLCc_GDPD_SF superfamily
7926 hypothetical protein
8369 hypothetical protein
8718 hypothetical protein
8737 hypothetical protein
9186 hypothetical protein
9339 hypothetical protein
9419 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007205 | protein kinase C activation | 0.00433615 | 1 | Thaps_bicluster_0175 |
| GO:0006725 | aromatic compound metabolism | 0.031426 | 1 | Thaps_bicluster_0175 |
| GO:0006413 | translational initiation | 0.0342405 | 1 | Thaps_bicluster_0175 |
| GO:0007165 | signal transduction | 0.0565048 | 1 | Thaps_bicluster_0175 |
| GO:0007242 | intracellular signaling cascade | 0.0592567 | 1 | Thaps_bicluster_0175 |
| GO:0006457 | protein folding | 0.173725 | 1 | Thaps_bicluster_0175 |
| GO:0006508 | proteolysis and peptidolysis | 0.349482 | 1 | Thaps_bicluster_0175 |


Comments